Biological and clinical significance of hepatitis B virus RNA splicing: an update
Introduction
Hepatitis B virus (HBV) is a hepadnavirus with a 3.2-kb, circular, partially double stranded DNA genome which is converted into covalently closed circular DNA (cccDNA) in the nucleus of infected hepatocytes. HBV genome contains four overlapping open reading frames (ORFs) encoding the hepatitis B e-antigen (HBeAg), the core protein, the reverse transcriptase-polymerase (P) protein, the X protein, and the large, middle and small surface proteins. Unidirectional transcription produces the pregenomic/precore mRNA and three subgenomic RNAs. The 3.5-kb bicistronic pregenomic RNA (pgRNA) serves as encapsidated template for reverse transcription during genome replication and as mRNA for translation of the core and the P protein. The precore mRNA is slightly longer and encodes the HBeAg. Subgenomic 2.4-kb preS1 and 2.1-kb preS2/S mRNAs encode the three surface proteins, and the small 0.7-kb mRNA encodes the X protein (1).
HBV cccDNA is transcribed by the cellular RNA polymerase II which is known to co-transcriptionally mediate splicing, capping, and polyadenylation of mRNAs. Consequently, it is assumed that a Cap at the 5' end modifies all viral RNAs. Although HBV RNAs are transcribed under the control of several distinct promoters (preC/pregenomic or basal core, S1, S2 and X promoters), they all terminate at a common polyadenylation signal (polyA). This particular feature generates mature mRNAs with long untranslated 3’ end sequences. In addition, singly and multiply spliced HBV transcripts from pgRNA and preS2/S mRNA have been identified from HBV-infected human liver tissues and serum, HBV-transgenic mice, and HBV-transfected hepatoma cell lines (2). Up to 80% of intracellular core particles and 20% of extracellular viral particles were reported to contain spliced genome in vitro (3).
Long-lasting persistence of HBV, also designed occult HBV infection/carriage (OBI), is characterized by the persistence of cccDNA in the nucleus of infected hepatocytes and very low levels of HBV DNA in plasma and/or in liver, with undetectable hepatitis B surface antigen (HBsAg) using the most sensitive commercial assays, with or without antibodies to hepatitis B core antigen (anti-HBc) or hepatitis B surface antigen (anti-HBs). It is associated with a strong inhibition of HBV replication and viral protein synthesis involving the complex interplay of still largely unclear viral and/or cellular control mechanisms of the viral genome expression. OBI may induce a mild continuous necroinflammation of the liver that may favor the development of cirrhosis and hepatocellular carcinoma (HCC) (4). Thus, the characterization of viral RNA metabolism including processing, maturation, nuclear export and stabilization/degradation of functional pregenomic and subgenomic HBV RNAs should provide insights into some of the molecular mechanisms essential for viral replication and potential valuable information for the development of new antiviral strategies.
The last data available in the literature were added to the HBV RNA splicing biological knowledge previously reviewed (5).
HBV RNA splicing
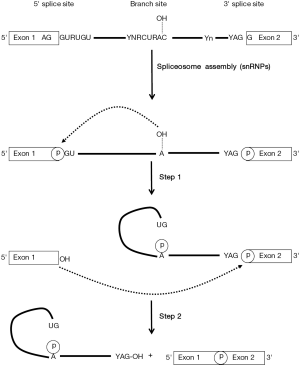
Splicing is needed to generate most of the functional eukaryotic mRNAs in which introns are removed and exons are joined precisely. This maturation process takes place in the nucleus after or concurrently with transcription and is mediated by a multiple heterogeneous nuclear ribonucleoproteins (hnRNPs)/RNA complex referred to as the spliceosome that assembles in a stepwise manner at the splice sites. The basic sequence elements located within the intron and required for splicing are a donor 5' splice site (5'ss), a branch point site including a critical adenine nucleotide, and an acceptor 3' splice site (3'ss) which usage is influenced by a polypyrimidine tract located closely upstream (Figure 1). Consensus splicing signal sequences have been established from the analysis of eukaryotic cellular genes and computer programs have been developed to identify putative splice-site locations in a given sequence (6,7). However, the possibility of site mis-identification remains an important issue for the prediction of splice sites. Despite these limitations, similar constitutive and cryptic splicing signals are predicted in the HBV genome in number and location that varies between genotypes and individual strain sequences.
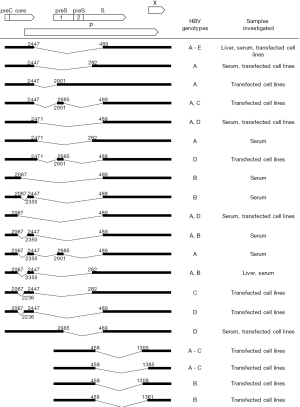
Based on experimental evidence, 16 splice variants of the pgRNA (3,8-15) and four splice variants of the preS2/S mRNA (16,17) have been identified so far (Figure 2). Single-, double- or rare triple-splice variants were characterized mainly in in vitro transfected cell experiments but also directly in liver tissues and sera of individuals infected with different HBV genotypes. Of note, all corresponding donor and acceptor sites reported in Figure 2, except 3'ss at position 2350, were correctly identified in consensus sequences of genotypes A-D by predictive programs (not shown). Different types of spliced variants derived from pgRNA or preS2/S RNA were frequently simultaneously detected in patient samples and in transfected cells indicating the existence of a complex alternative splicing pattern of both HBV pregenomic and subgenomic RNAs. The most frequently reported and studied pgRNA splice variants are two 2.2-kb spliced isoforms: a singly spliced product 1 (SP1) between nucleotides 2447 and 489, and the doubly spiced product between nucleotides 2447 and 2901 and between nucleotides 2985 and 489 (13). The SP1 splice variant was found to account for 25–30% of the pgRNA in transfected hepatoma-derived cell cultures and for 50–60% of all splice variants detected in patient samples (3,11,13,15).
Efficiency of pgRNA splicing and nature of splice variants differ from patient to patient ranging from undetectable splicing to extensive and genotype/viral strain-dependent splicing (3,9,11,13,15). One possible reason for such variation may be that less prevalent splice variants are not detected due to insufficient sensitivity of RNA amplification-based detection methods. Unspliced RNA and spliced variant(s) are usually co-amplified in a single competitive RT-PCR reaction and significant size difference may affect final amplification efficiency. The specificity and location of the primer combinations used in different studies may also affect detection. Amplification of the entire viral mRNA investigated should be preferred to subgenomic amplifications. Inter-genotypes and inter-patients sequence variations may also create, disrupt or modulate the function of splicing signal sequences with qualitative and quantitative effects on HBV splicing (18). Furthermore, some studies identified HBV RNA splicing based on the detection of defective genomes isolated from viral particle present in patient sera or cell culture medium. In this case, only splice variants retaining regulatory sequences required for RNA encapsidation and reverse transcription could be detected. Therefore, the overall extent of splicing might be underestimated as some splice variants would be present only in the cytoplasm of infected or transfected cells (e.g., preS2/S splice variants). The popular utilization of human hepatoma cell lines Huh7 and HepG2 transfected with plasmids containing fully replicative or partial HBV genome may introduced some bias too. Increase in splicing was observed when using the cytomegalovirus immediate early (CMV-IE) promoter to drive transcription of viral pgRNA or subgenomic RNAs compared to endogenous HBV promoters, and HepG2 cells produced a higher level of spliced RNAs as compared to Huh7 cells irrespective of the promoter used (14; personal observations).
Regulation of HBV RNA splicing
The different levels and patterns of splicing observed in transfected cell cultures, patient samples and different genotype or viral strain sequences indicate that HBV RNA splicing is not a constitutive process. The comparison of the actually used splicing sites with the predicted splicing sites shows that some of the used splice sites are not necessarily predicted (3'ss position 2350 in Figure 2 for example) and some predicted ones are apparently not used. Interestingly, the 5'ss position 458 and the 3'ss positions 1305 and 1385 generating the major pre-S2/S spliced variants are also present in the pgRNA sequence. However, 458–1305/85 spliced variants of the pgRNA or the corresponding encapsidated defective DNA have never been described despite the putative spliced RNAs that would retain functional encapsidation signals. Furthermore, in most eukaryote cells, pre-mRNA splicing is closely linked to mRNAs export out of the nucleus via the recruitment of specific export factors through direct interaction with spliceosomal proteins, and unspliced RNA is generally rapidly hydrolyzed in the nucleus. This creates a paradox for nucleocytoplasmic transport of partly spliced and unspliced HBV RNA. It is critical for the viral life cycle that the majority of viral RNAs escapes the cellular splicing machinery and is exported by a splicing-independent mechanism. This suggests that HBV splicing is precisely regulated by cis-acting genetic elements and cellular and/or viral trans-acting factors controlling both activation of cryptic splicing sites and silencing of constitutive splicing sites.
Accurate alternative splicing does not only require the recognition of 5'ss, 3'ss and branch point sequence. Exonic and intronic cis-regulatory elements [splicing regulatory elements (SREs)] are required to recruit sequence-specific RNA-binding protein factors that activate or repress the utilization of adjacent splicing sites (19). These elements are conventionally classified as exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs), intronic splicing enhancers (ISEs), and intronic splicing silencers (ISSs) according to their location and function. Usually, splicing enhancers tend to bind serine/arginine-rich (SR) proteins and splicing silencers to recruit hnRNPs.
A cis-acting regulatory element known as the post-transcriptional regulatory element (PRE) was identified within the HBV genome at nucleotides 1217–1582. This PRE is present within the 3' end of all HBV mRNAs. Several studies showed that PRE serves as a crucial orientation-dependent nuclear export element for unspliced preS/S mRNAs by recruiting the TREX complex via a 116 nucleotides functional sub-element (SEP1) and also contributed to pgRNA stability and possibly 3' end processing, but without significant influence on its nuclear export (2,20,21). HBV PRE appears to be highly conserved among HBV genotypes and contains two conserved secondary structured elements (stem-loops) termed HBV SLα and HBV SLβ the function of which has not yet been clearly established (22). However, the 3'ss at position 1305 is embedded within SLα and it has been suggested that the secondary structure of this element may prevent the use of this splicing site to allow the nuclear export of unspliced HBV RNAs (23). Pre-mRNA secondary structures have been reported to influence splicing through occlusion/exposure of splicing sites or cis-acting regulatory elements and spatial modification of the distance between these elements (24). For instance, regulation of the related duck hepatitis virus (DHBV) pgRNA splicing depends on a secondary structure of the viral RNA that contains the 5'ss and 3'ss and brings them in close proximity (25).
Further functional analysis identified within the PRE distinct elements that positively and negatively regulate pgRNA splicing in transfected cultured cells. A splicing enhancer element termed splicing regulatory element 1 (SRE1) was characterized upstream to the PRE SLα and SLβ domains (nucleotides 1252–1288) (26), and a splicing silencer was located at the 3' region of PRE (nucleotides 1481–1585) (27). This silencer element folds into a double-hairpin structure and both sense and antisense strands strongly suppress alternative splicing in a sequence-independent manner. Other potential cis-acting regulatory elements have been located upstream of PRE. Zu Putlitz et al. have shown that a 30-nucleotides deletion between 5'ss at position 458 and 3'ss at position 489 significantly reduced the expression of preS/S RNAs (28). In addition, both splicing activator (nucleotides 2951–2970 and 3051–3070) and inhibitor (nucleotides 3138–3143) elements have been reported within an intronic splicing regulator domain in pgRNA (29). However, these data have been generated with very artificial in vitro systems and the function of these elements in the natural regulation of HBV splicing remains elusive.
It is assumed that the activity of splicing regulatory elements is regulated by trans-acting factors that are still largely unknown. Cellular splicing regulatory proteins (SF2/ASF, SC35, SRp45 and SRp55) are predicted to bind to SRE1 but this was not confirmed by experimental data (2). However, the PTB-associated splicing factor (PSF) was found to strongly stimulate pgRNA splicing most likely by interacting directly with the viral RNA, especially in the absence of PRE (26). PSF interacts with the polypyrimidine tract close to the 3'ss and may replace the 3' splice site binding factor U2AF65 during the second catalytic step of splicing (30). In addition, several studies showed that another splicing regulatory protein, the polypyrimidine tract binding protein (PTB), interacts with the 3'-end of the PRE, represses pgRNA splicing and facilitates unspliced preS/S RNA nuclear export (26,31,32). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was also reported to bind to the PRE but the functional meaning of such binding remains unknown (33).
There are several lines of evidence suggesting that HBV core protein may be involved in RNA splicing. The core proteins cross the nuclear envelop in infected cells and interact with pgRNA during encapsidation (34,35). The carboxy-terminal sequence of the core protein constitutes an unspecific DNA and RNA-binding domain that is rich in arginine residues and contains three serine phosphorylation sites. Mutations in this carboxy-terminal sequence resulted in a reduced level of spliced RNA that was preferentially encapsidated (36,37). Furthermore, a correlation between preferential nuclear localization of the core protein and occurrence of viral RNA splicing has been reported (38). Interestingly, a minor fraction of the DHBV core protein was also reported to accumulate in nuclear subdomains close to splicing factor compartments (39). Taken together, these findings suggest that the hepadnavirus core protein may have a nonstructural function in the posttranscriptional viral RNA metabolism.
Novel proteins encoded by HBV spliced RNAs
So far, three proteins translated from spliced viral RNAs have been partially characterized. The 43 kDa polymerase-surface fusion glycoprotein (P-S FP) generated by 2447–2901 splicing has been detected in cell lysates, virions and subviral particles (40). The P-S FP was not needed for in vitro viral replication but it was able to substitute for the HBV long surface protein in virion maturation. It was speculated that this protein may function as a structural protein involved in viral entry. Another study reported that P-S FP localized in the endoplasmic reticulum and inhibited HBV replication in transfected cells through a mechanism remaining to be characterized (41). Interestingly, the similar L protein encoded by a spliced RNA derived from the DHBV pgRNA was found functionally important for virus replication in infected duck hepatocytes, but not for virus formation from transfected DNA genomes, and may play a distinct role in an early step of the viral life cycle (42).
The hepatitis B doubly spliced protein (HBDSP) is generated by the 2.2-kb doubly spliced pgRNA frequently detected in patients with chronic hepatitis B and in various transfected hepatocyte-derived cell lines (43). It has been suggested that HBDSP could act as a pleiotropic activator protein with modest transactivating activities mediated by activator protein-1- and CCAAT/enhancer-binding protein-binding sites. However, the exact mechanisms underlying the transactivational activities of HBDSP, its biological significance, and potential role in HBV pathogenesis remain unclear.
The SP1 variant encodes the third known fusion protein called the HBV splice-generated protein (HBSP) (44). This 93 amino acids protein derives from the fusion of the first 46 amino acids of the polymerase and 47 amino acids of a new ORF that is created by the splicing event. This protein has been found to be expressed in liver tissues of patients with chronic HBV infection. Antibodies to HBSP and HBSP-specific T-cell responses have been identified in HBV-infected patients but no prominent role in liver disease was observed (44,45). It has been suggested that the relative immunodominance of HBSP compared to HBV structural proteins may provide an immune decoy-based mechanism for viral persistence in chronic infection (46). The ectopic expression of HBSP showed no effect on viral DNA replication or transcription but caused cell apoptosis in vitro (44,47). The presence of antibodies to HBSP in HBV chronic carriers has also been associated with viral replication, elevation of tumor necrosis factor alpha (TNFα), and severity of liver fibrosis (48). However, another study suggested that HBSP could bind to fibrinogen gamma chain and inhibit fibrin polymerization (49). No cytopathic effect of HBSP in vivo has been shown and its possible role in progression, invasion, and metastasis of HBV-related HCC has not yet been elucidated. Interestingly, in vitro HBSP overexpression and interaction with cathepsin B enhanced tumor-induced angiogenesis, appeared to activate the mitogen-activated protein kinase and Akt signaling pathway, and may promote hepatoma cell mobility and invasion, thereby contributing to the aggressiveness of hepatoma cells (50). Recently, HBSP has been reported to hack TNF-α-stimulated signaling pathways involved in innate immunity and to limit the extent of liver inflammation in mice (51). Similarly, an elevation of the level of HBV spliced variants in the sera of patients with chronic HBV infection has been reported to correlate with an impaired response to interferon-α treatment. Expression of the HBSP and the N-terminal-truncated viral polymerase protein resulted in a strong suppression of IFN-α signaling transduction in both cell and mouse models (52). Further studies are still required to determine if these alternative-splicing regulated viral proteins are part of an additional strategy developed by the virus to escape innate and/or adaptive immunity.
A 21.5 kDa viral protein (p21.5) encoded by the 2.2 kb singly-spliced RNA (nt2447–nt489) was detected in hepatoma cells (53). Expression of this protein, one-amino acid shorter than the precore/core protein, has been reported to interfere with nucleocapsid formation, raising the possibility that the elevation of singly-spliced RNA in chronic infected patients may play a functional role in the feedback modulation of HBV replication. In this case, the increased level of p21.5 translated from the singly-spliced RNA may decrease the formation of nucleocapsid both in vitro and in vivo and thus may contribute to the establishment of a viral persistence.
The detection of proteins encoded by several spliced RNAs advocates for a potential function of viral RNA alternative splicing during HBV infection. More investigations are needed to identify other putative new viral proteins and clarify the function of the few already known. However, it might be of interest to pay attention not only to the encoding capacity of rearranged exons but also to introns as proper complex entities and not just splicing waste products. Recent improvements in high-throughput gene expression analysis revealed that noncoding RNAs (ncRNAs) represent the large majority of the human transcriptome and their regulatory role in gene expression is now broadly recognized. Interestingly, non-degradated long intronic ncRNAs have been found to be selectively exported to the cytoplasm and involved in global gene expression regulation, either as direct trans-acting regulatory elements or as precursors of shorter regulatory microRNAs (miRNAs) [(54) for review]. Reciprocal interactions and cross-talk between viral coding/non coding RNAs and host mRNAs and miRNAs have been reported in the literature (55). In HBV RNAs, several sites that are complementary to host miRNAs (e.g., miR-122, miR-15a/16 cluster) involved in the regulation of viral gene expression, viral replication and tumor suppression have been identified [(55) for review]. These complementary sites acting as sponges to bind and sequester host miRNAs may favor persistent viral infection and HCC development. Interestingly, a recent study showed that the expression of 2.2 double-spliced RNA reduced significantly the level of HBV pgRNA and X RNA in transfected human hepatoma cells and in HBV transgenic mice (56). The 2.2 double-spliced RNA acted as a repressor of viral transcription through an interaction with the TATA-binding protein (TBP) that induced stress granule formation. These data suggested that a spliced RNA may act as a suppressive noncoding RNA that regulating HBV replication. Additional studies are required to determine if other HBV spliced RNAs and/or intronic ncRNAs may be involved in RNA-based regulatory processes and the development of chronic hepatitis B.
Splicing during the course of HBV infection
A function for spliced RNAs and the corresponding putative encoded proteins is strongly supported by the high prevalence of spliced variants in HBV-infected patients [~50% in asymptomatic HBsAg+ blood donors (15,57) to >90% in patients with mild active chronic hepatitis (12) or HCC (58)] and the sequence conservation of splicing sites in HBV sequences irrespective of genotype. RNA splicing has also been described in duck and woodchuck hepatitis viruses suggesting some strong functional constraints to maintain splicing ability along hepadnavirus evolution (25,42,59). However, splicing results in the disruption of the HBV ORFs coding for the structural viral proteins and mutagenesis of the splice sites showed that pgRNA splicing is not required for viral replication in vitro (9,11).
Encapsidation of spliced pgRNAs results in the production of viral particle containing a defective viral DNA genome (3,10,12,13,15). The co-expression of spliced and unspliced genomes in the same hepatocyte is necessary for the trans-complementation and release of spliced variants from infected cells. In co-transfection experiments, viral proteins produced from a full-length viral genome have been shown to enhance the replication of a pgRNA double-spliced variant without necessary replication of the unspliced genome (58). Certain defective HBV genome variants were reported to behave like defective interfering particles that contributed to viral persistence by attenuating wild-type virus replication as described in other viruses (56,60). In contrast, another study reported that a 2.2-kb double-spliced variant enhanced the replication of its matched 3.2-kb full-length HBV genome and other heterologous full-length genomes from genotypes B and C isolates when co-transfected into HepG2 cells (58).
The sequential detection of spliced viral DNA in seroconversion panels showed that splicing coincided with or shortly followed HBsAg detection during the early phase of HBV infection in only some individuals (15). In another study, preS2/S mRNA splicing seemed to be essential for the production of detectable level of HBsAg (16). These data suggest that, compared to the highly infectious preseroconversion window period, HBsAg detectability and the concomitant lower infectivity might be related to the initiation of pgRNA and/or preS2/S mRNA splicing. The existence and extent of circulating spliced variants should be considered when determining the HBV infectious dose (15).
Spliced variants were also found predominantly in patients chronically infected HBsAg+ and rarely in those with acute self-limited clinical course (3,10,12,13). However, these observations should be considered with caution since HBV splicing detection efficacy may vary according to the nature of the viral nucleic acid tested (viral RNA or DNA in infected hepatocytes versus circulating encapsidated DNA in serum) and to the performance of the molecular assays used as mentioned above. In addition, the relative amount of spliced variants differs from patient to patient and between different genotypes (3,15,57) and sequential studies of individual patients using standardized assays may better characterize the clinical significance of these spliced variants. Nevertheless, the expression of the SP1 variant within liver biopsies was reported to correlate with a low histology activity index (38) and decreased HBV RNA splicing has been associated with complex HBV variants and progression into cirrhosis (61). Other studies showed that the expression of the 2.2-kb spliced DNA in transfected Huh7 cells resulted in a significant nuclear and cytoplasmic accumulation of the core protein and an increased secretion of HBeAg that may favor immunotolerance (12,38,44). These observations support the notion that HBV RNA splicing may occur preferentially in the early phases of chronic HBV infection and the hypothesis that an immunomodulatory function of the splice variants may contribute to HBV persistence.
In contrast, other investigators reported the presence of spliced variants in tumor tissues of patients with HCC (62). The relative content of a 2.2-kb spliced variant was reported to be higher in tumor tissues than that in the peri-tumor tissues (62). In another study, the mean ratio of spliced HBV DNA to total (spliced plus unspliced) HBV DNA was higher in patients with severe liver necrosis and fibrosis (63). These observations suggest a possible role of the spliced variants in the pathogenesis of HCC development. The 2.2-kg spliced variant has been shown to enhance the replication of full-length HBV genome in cell cultures and these enhancing effects required the expression of the X gene (58). The X protein is known as a transactivating protein that can activate viral genes and oncogenes (64). In theory, all described spliced variants, except preS2/S variants, retain potentially functional enhancer I/X promoter and X gene sequences. It has been hypothesized that the enhancing replicative effect of the X gene carried by spliced variants may play a role in persistent HBV replication in a subpopulation of HCC patients (58).
Recently, it has been reported that following the integration of HBV into the intron of the cyclin A2 gene, the mammalian splicing machinery utilized the foreign splice sites at nt282 and nt458 of the HBV genome to generate a pseudo-exon, forming an in-frame HBV-cyclin A2 chimeric transcript, as detected in 12.5% of HCC patients, but in none of the non-HCC HBV-associated cirrhotic liver samples examined (65). The fusion chimeric protein gained a non-degradable property and promoted cell cycle progression demonstrating its potential oncogenic functions. The generation by the mammalian pre-mRNA splicing machinery of HBV-human chimeric transcripts from intronically integrated HBV genome may have a potential tumorigenic impact and participate to the development of HBV-related HCC.
HBV splicing and occult infection
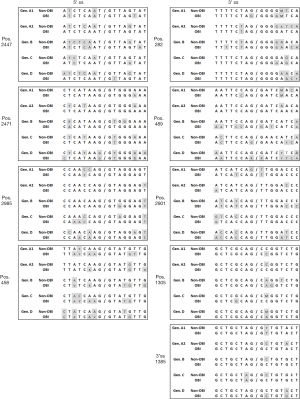
Splicing of HBV pgRNA has been extensively investigated in HBsAg positive chronically infected individuals and correlated with the overall viral load (15). It is tempting to speculate that this mechanism may be involved in some specific aspects of OBI like the apparent limited viral replication. However, there is no data available about the frequency of pgRNA splicing in OBI. The HBV DNA load in the blood of OBI carriers is generally extremely low and the standard amplification methods may lack the sensitivity required for appropriate detection of encapsidated unspliced and spliced genomes. In our experience, very few full genomes can be obtained and only partial genome sequences (400–1200 nucleotides) are successfully amplified in the majority of OBI samples (66). They are of limited interest to assess the replicative properties of HBV strains in in vitro transfection experiments. In addition, liver biopsies from OBI carriers presenting no sign of liver disease are not available for investigations due to ethical reasons. Generally, the commonly recognized minimal pgRNA splicing sites appear conserved in OBI sequences compared to non-OBI sequences, suggesting that in the majority of cases OBI viral RNAs may retain the ability to undergo splicing irrespective of genotypes (Figure 3). However, sequence variability in some genotypes at some particular splicing sites might affect splicing: 5'ss position 2471 and 3'ss position 489 in OBI genotype D sequences, 3'ss position 489 in OBI genotype B sequences, and 3'ss position 2901 in both OBI and non-OBI genotype A1 sequences (Figure 3). Sequence variations are mainly observed in the vicinity of the canonical splicing sites but without a clear association with the OBI status, except for OBI sequences close to the 5'ss position 458 (Figure 3). RNA secondary structures are essential for appropriately displaying splicing sequences and recruiting splicing proteins (67). Considering that there are as many examples of RNA structures aiding splicing as there are structures inhibiting splicing, specific mutations may induce changes in RNA structures that favor different splicing outcomes. In addition, the possibility that the high genetic variability usually observed across the OBI genome might negatively affect some splicing regulatory elements cannot be excluded (17).
Aiming to investigate the functional role of HBV mutations in OBI carriers experiencing viral reactivation, a single mutation affecting HBsAg expression by a post-transcriptional mechanism was described (16). Sequence analysis and site directed mutagenesis (SDM) showed that a single G→A substitution (G458A) at the 5'ss position 458 in the S gene induced downregulation of HBsAg expression and inhibited preS2/S mRNA splicing. Paradoxically, inhibition of splicing resulted in a marked decrease of the functional preS2/S mRNA and its translation product. The authors hypothesized that the G458A mutation may alter interaction between preS2/S RNA and exon junction complex proteins by disrupting the RNA putative secondary structure at the 5'ss (Figure 4), thus modulating post-transcriptional RNA processing. RNA structures have been identified that inhibit splicing by sequestering 5'ss, 3'ss or branch-point in base-paired structures similar to the one predicted at the native HBV preS2/S 5'ss (67). However, this structure was predicted using the computer modeling program MFOLD and has not yet been validated by using biochemical methods or by testing compensatory mutations to restore the proposed structure. Another issue to keep in mind is the fact that splicing can occur co-transcriptionally before slow folding RNA structures might form and the possible competition between RNA folding and protein binding to single-stranded RNA conformations (67).
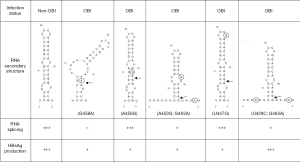
While the G458A mutation has not yet been reported in another OBI viral strain, a significantly higher frequency of mutations in the vicinity of the pre-S2/S 5'ss at position 458 is observed in OBI than in non-OBI strains while the 3'ss at positions 1305 and 1385 are mainly conserved in both type of strains, irrespective of HBV genotype (Figure 3). It has been recently reported that 25/55 (45%) genotype B OBI and 14/33 (42%) genotype C OBI sequences from blood donors presented mutations in the vicinity of the 5'ss 458, compared with 5/47 (11%) and 5/48 (10%) of genotype B and C non-OBI sequences, respectively (17). Genotypes B and C non-OBI sequences were predicted to fold in the same stem-loop secondary structure previously described (Figure 4) (16,17). Among OBI variants, 44% of genotype B OBI and 36% of genotype C OBI heterogeneous splice donor mutations disrupted this stem-loop structure as described with the G458A mutation. Similar disrupting mutations have been also recently reported in 4.5% of HBV sequences obtained from OBI patients (68). None of the few substitutions found in non-OBI variants was predictive of affecting the stem-loop structure. Results of transient transfection of Huh7 cells with plasmids expressing the S-coding region from a limited number of OBI strains and non-OBI controls suggested that some disruptive mutations were indeed associated with decreased splicing efficiency, but it did not correlate with the level of HBsAg secretion (17). It was hypothesized that the lack of correlation between the apparent yields of amplified spliced RNAs and HBsAg production may be related to more efficient amplification of the shorter spliced RNA than unspliced RNA. Nevertheless, additional studies are needed to investigate the effect of mutations impairing the usage of preS2/S 3'ss and/or the corresponding branch-point sites to further support a direct functional association between splicing and HBsAg production.
It is still unclear whether mutations potentially affecting the structure of the preS2/S 5'ss or splicing itself directly negatively influence HBsAg production, and further investigations from a larger number of OBI strains are needed. However, there are some lines of evidence strongly suggesting that the 5'ss 458 is part of a regulatory element essential for maintaining high preS2/S transcript levels and HBsAg production. The process of splicing itself might not be directly involved but RNA splicing is also known to be coupled with RNA nuclear export. Therefore, it can be hypothesized that mutations, including G458A, within such a putative regulatory element may affect RNA-proteins interactions upon formation of the spliceosome thus resulting in reduced nuclear export or RNA stability. However, the posttranscriptional mechanism of HBsAg expression downregulation and its responsibility in the lack of HBsAg detection characterizing occult HBV carriage remain non-elucidated (17,69).
Perspectives
Splicing of HBV RNAs is firmly established and adds another level of complexity in the expression of a relatively small viral genome. The high conservation of splicing sites, the complex modulation of splicing during the course of infection, and the presence of unique proteins encoded by spliced viral RNAs are strongly suggestive of a functional role of splicing in the HBV life cycle. Additional studies are required to characterize host factors, additional cis-acting elements, RNA-RNA, RNA-protein and protein-protein interactions involved in the complex regulatory mechanisms underlying HBV RNA splicing. Understanding the regulation of HBV splicing might provide valuable indications on its potential implication in the long-term persistence of HBV in occult carriers.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aob.2017.05.01). DC serves as an unpaid editorial board member of Annals of Blood from Dec 2016 to Dec 2019. JPA serves as an unpaid editorial board member of Annals of Blood from Dec 2016 to Dec 2018. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev 2000;64:51-68. [Crossref] [PubMed]
- Sommer G, Heise T. Posttranscriptionnal control of HBV gene expression. Front Biosci 2008;13:5533-47. [Crossref] [PubMed]
- Sommer G, van Bömmel F, Will H. Genotype-specific synthesis and secretion of spliced hepatitis B virus genomes in hepatoma cells. Virology 2000;271:371-81. [Crossref] [PubMed]
- Pollicino T, Saitta C. Occult hepatitis B virus and hepatocellular carcinoma. World J Gastroenterol 2014;20:5951-61. [Crossref] [PubMed]
- Candotti D, Allain JP. Biological and clinical significance of hepatitis B virus RNA splicing: a role for spliced variants in occult infection?. In: Allain JP, Fu Y, Li C, et al. editors. Occult hepatitis B infection, 1st ed. Beijing: Science Press, 2015:90-107.
- Wang M, Marin A. Characterization and prediction of alternative splice sites. Gene 2006;366:219-27. [Crossref] [PubMed]
- Dogan RI, Getoor L, Wilbur WJ, et al. SplicePort-An interactive splice-site analysis tool. Nucleic Acids Res 2007;35:W285-91 [Crossref] [PubMed]
- Chen PJ, Chen CR, Stung JL, et al. Identification of a double spliced viral transcript joining the separated domains for putative protease and reverse transcriptase of hepatitis B virus. J Virol 1989;63:4165-71. [PubMed]
- Su TS, Lai CJ, Huang JL, et al. Hepatitis B virus transcript produced by RNA splicing. J Virol 1989;63:4011-8. [PubMed]
- Terré S, Petit AM, Bréchot C. Defective hepatitis B virus particles are generated by packaging and reverse transcription of spliced viral RNAs in vivo. J Virol 1991;65:5539-43. [PubMed]
- Wu HL, Chen PJ, Tu SJ, et al. Characterization and genetic analysis of alternatively spliced transcripts of hepatitis B virus in infected human liver tissues and transfected HepG2 cells. J Virol 1991;65:1680-6. [PubMed]
- Rosmorduc O, Petit AM, Pol S, et al. In vivo and in vitro expression of defective hepatitis B virus particles generated by spliced hepatitis B virus RNA. Hepatology 1995;22:10-9. [PubMed]
- Günther S, Sommer G, Iwanska A, et al. Heterogeneity and common features of defective hepatitis B virus genomes derived from spliced pregenomic RNA. Virology 1997;238:363-71. [Crossref] [PubMed]
- Abraham TM, Lewellyn EB, Haines KM, et al. Characterization of the contribution of spliced RNAs of hepatitis B virus to DNA synthesis in transfected cultures of Huh7 and HepG2 cells. Virology 2008;379:30-7. [Crossref] [PubMed]
- El Chaar M, El Jisr T, Allain JP. Hepatitis B virus DNA splicing in Lebanese blood donors and genotype A to E strains: implication for hepatitis B virus DNA quantification and infectivity. J Clin Microbiol 2012;50:3159-67. [Crossref] [PubMed]
- Hass M, Hannoun C, Kalinina T, et al. Functional analysis of hepatitis B virus reactivating in hepatitis B surface antigen-negative individuals. Hepatology 2005;42:93-103. [Crossref] [PubMed]
- Candotti D, Lin CK, Belkhiri D, et al. Occult hepatitis B infection in blood donors from South East Asia: molecular characterization and potential mechanisms of occurrence. Gut 2012;61:1744-53. [Crossref] [PubMed]
- Lewandowska MA. The missing puzzle piece: splicing mutations. Int J Clin Exp Pathol 2013;6:2675-82. [PubMed]
- Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 2008;14:802-13. [Crossref] [PubMed]
- Guang S, Mertz JE. Pre-mRNA processing enhancer (PPE) elements from intronless genes play additional roles in mRNA biogenesis than do ones from intron-containing genes. Nucl Acids Res 2005;33:2215-26. [Crossref] [PubMed]
- Chi B, Wang K, Du Y, et al. A sub-element in PRE enhances nuclear export of intronless mRNAs by recruiting the TREX complex via ZC3H18. Nucl Acids Res 2014;42:7305-18. [Crossref] [PubMed]
- Smith GJ, Donello JE, Lück R, et al. The hepatitis B virus post-transcriptional regulatory element contains two conserved RNA stem-loops which are required for function. Nucleic Acids Res 1998;26:4818-27. [Crossref] [PubMed]
- Schwalbe M, Ohlenschläger O, Marchanka A, et al. Solution structure of stem-loop alpha of the hepatitis B virus post-transcriptional regulatory element. Nucleic Acids Res 2008;36:1681-9. [Crossref] [PubMed]
- Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol 2004;24:10505-14. [Crossref] [PubMed]
- Loeb DD, Mack AA, Tian R. A secondary structure that contains the 5’ and 3’ splice sites suppresses splicing of duck hepatitis B virus pregenomic RNA. J Virol 2002;76:10195-202. [Crossref] [PubMed]
- Heise T, Sommer G, Reumann K, et al. The hepatitis B virus PRE contains a splicing regulatory element. Nucleic Acids Res 2006;34:353-63. [Crossref] [PubMed]
- Huang C, Xie MH, Liu W, et al. A structured RNA in hepatitis B virus post-transcriptional regulatory element represses alternative splicing in a sequence-independent and position-dependent manner. FEBS J 2011;278:1533-46. [Crossref] [PubMed]
- zu Putlitz J, Tong S, Wands JR. A short region in the genome of hepatitis B virus is critical for maintenance of high transcript levels. Virology 1999;254:245-56. [Crossref] [PubMed]
- Chowdhury JB, Roy D, Ghosh S. Identification of a unique splicing regulatory cluster in hepatitis B virus pregenomic RNA. FEBS Lett 2011;585:3348-53. [Crossref] [PubMed]
- Gozani O, Patton JG, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J 1994;13:3356-67. [PubMed]
- Zang WQ, Li B, Huang PY, et al. Role of polypyrimidine tract binding protein in the function of the hepatitis B virus posttranscriptional regulatory element. J Virol 2001;75:10779-86. [Crossref] [PubMed]
- Roy D, Chowdhury JB, Ghosh S. Polypyrimidine tract binding protein (PTB) associates with intronic and exonic domains to squelch nuclear export of unspliced RNA. FEBS Lett 2013;587:3802-7. [Crossref] [PubMed]
- Zang WQ, Fieno AM, Grant RA, et al. Identification of glyceraldehyde-3-phosphate dehydrogenase as a cellular protein that binds to the hepatitis b virus posttranscriptional regulatory element. Virology 1998;248:46-52. [Crossref] [PubMed]
- Beames B, Lanford RE. Carboxy_terminal truncations of the HBV core protein affect capsid formation and the apparent size of encapsidated HBV RNA. Virology 1993;194:597-607. [Crossref] [PubMed]
- Li HC, Huang EY, Su PY, et al. Nuclear export and import of human hepatitis B virus capsid protein and particles. PLoS Pathog 2010;6:e1001162 [Crossref] [PubMed]
- Köck J, Nassal M, Deres K, et al. Hepatitis B virus nucleocapsids formed by carboxy-terminally mutated core proteins contains spliced viral genomes but lack full-size DNA. J Virol 2004;78:13812-8. [Crossref] [PubMed]
- Le Pogam S, Chua PK, Newman M, et al. Exposure of RNA templates and encapsidation of spliced viral RNA are influenced by the arginine-rich domain of human hepatitis B virus core antigen (HBcAg 165-173). J Virol 2005;79:1871-87. [Crossref] [PubMed]
- Sheen IS, Tsou YK, Lin SM, et al. Nuclear HBcAg and histology activity index as independent predictors of the expression of singly spliced HBV-RNA. J Viral Hepat 2007;14:70-4. [Crossref] [PubMed]
- Mabit H, Knaust A, Breiner KM, et al. Nuclear localization of the duck hepatitis B virus capsid protein: detection and functional implications of distinct subnuclear bodies in a compartment associated with RNA synthesis and maturation. J Virol 2003;77:2157-64. [Crossref] [PubMed]
- Huang HL, Jeng KS, Hu CP, et al. Identification and characterization of a structural protein of hepatitis B virus: a polymerase and surface fusion protein encoded by a spliced RNA. Virology 2000;275:398-410. [Crossref] [PubMed]
- Park GS, Kim HY, Shin HS, et al. Modulation of hepatitis B virus replication by expression of polymerase-surface fusion protein through splicing: implications for viral persistence. Virus Res 2008;136:166-74. [Crossref] [PubMed]
- Obert S, Zachmann-Brand B, Deindl E, et al. A spliced hepadnavirus RNA that is essential for virus replication. EMBO J 1996;15:2565-74. [PubMed]
- Chen WN, Chen JY, Lin WS, et al. Hepatitis B doubly spliced protein, generated by a 2.2 kb doubly spliced hepatitis B virus DNA, is a pleiotropic activator protein mediating its effect via activator protein-1- and CCAAT/enhancer-binding protein-binding sites. J Gen Virol 2010;91:2592-600. [Crossref] [PubMed]
- Soussan P, Garreau F, Zylberberg H, et al. In vivo expression of a new hepatitis B virus protein encoded by a spliced RNA. J Clin Invest 2000;105:55-60. [Crossref] [PubMed]
- Bayard F, Godon O, Nalpas B, et al. T-cell responses to hepatitis B splice-generated protein of hepatitis B virus and inflammatory cytokines/chemokines in chronic hepatitis B patients. ANRS study: HB EP 02 HBSP-FIBRO. J Viral Hepat 2012;19:872-80. [Crossref] [PubMed]
- Lee GH, Wasser S, Lim SG. Hepatitis B pregenomic RNA splicing-The products, the regulatory mechanisms and its biological significance. Virus Res 2008;136:1-7. [Crossref] [PubMed]
- Lu YW, Tan TL, Chan V, et al. The HBSP gene is expressed during HBV replication, and its coded BH3-containing spliced viral protein induces apoptosis in HepG2 cells. Biochem Biophys Res Commun 2006;351:64-70. [Crossref] [PubMed]
- Soussan P, Tuveri R, Nalpas B, et al. The expression of hepatitis B spliced protein (HBSP) encoded by a spliced hepatitis B virus RNA is associated with viral replication and liver fibrosis. J Hepatol 2003;38:343-8. [Crossref] [PubMed]
- Chen JY, Chen WN, Liu LL, et al. Hepatitis B spliced protein (HBSP) generated by a spliced hepatitis B virus RNA participates in abnormality of fibrin formation and functions by binding to fibrinogen gamma chain. J Med Virol 2010;82:2019-26. [Crossref] [PubMed]
- Chen WN, Chen JY, Jiao BY, et al. Interaction of the hepatitis B spliced protein with cathepsin B promotes hepatoma cell migration and invasion. J Virol 2012;86:13533-41. [Crossref] [PubMed]
- Pol JG, Lekbaby B, Redelsperger F, et al. Alternative splicing-regulated protein of hepatitis B virus hacks the TNF-α-stimulated signaling pathways and limits the extent of liver inflammation. FASEB J 2015;29:1879-89. [Crossref] [PubMed]
- Chen J, Wu M, Wang F, et al. Hepatitis B virus spliced variants are associated with an impaired response to interferon therapy. Sci Rep 2015;5:16459. [Crossref] [PubMed]
- Wang YL, Liou GG, Lin CH, et al. The inhibitory effect of the hepatitis B virus singly-spliced RNA-encoded p21.5 protein on HBV nucleocapside formation. PLoS One 2015;10:e0119625 [Crossref] [PubMed]
- Louro R, Smirnova AS, Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics 2009;93:291-8. [Crossref] [PubMed]
- Li C, Hu J, Hao J, et al. Competitive virus and host RNAs: the interplay of a hidden virus and host interaction. Protein Cell 2014;5:348-56. [Crossref] [PubMed]
- Tsai KN, Chong CL, Chou YC, et al. Doubly spliced RNA of hepatitis B virus suppresses viral transcription via TATA-binding protein and induces stress granule assembly. J Virol 2015;89:11406-19. [Crossref] [PubMed]
- Cox LE, Arslan O, Allain JP. Characterization of hepatitis B virus in Turkish blood donors, and the prevalence of the SP1 splice variant. J Med Virol 2011;83:1321-5. [Crossref] [PubMed]
- Ma ZM, Lin X, Wang YX, et al. A double-spliced defective hepatitis B virus genome derived from hepatocellular carcinoma tissue enhanced replication of full-length virus. J Med Virol 2009;81:230-7. [Crossref] [PubMed]
- Ogston CW, Razman DG. Spliced RNA of woodchuck hepatitis virus. Virology 1992;189:245-52. [Crossref] [PubMed]
- Yuan TT, Lin MH, Chen DS, et al. A defective interference-like phenomenon of human hepatitis B virus in chronic carriers. J Virol 1998;72:578-84. [PubMed]
- Märschenz S, Endres AS, Brinckmann A, et al. Functional analysis of complex hepatitis B virus variants associated with development of liver cirrhosis. Gastroenterology 2006;131:765-80. [Crossref] [PubMed]
- Lin X, Wen Y, Wan D, et al. Structural and functional analysis of 2.2 kb spliced variant of hepatitis B virus genomes isolated from liver tissues from hepatocellular carcinoma patients. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 2002;16:11-5. [PubMed]
- Soussan P, Pol J, Garreau F, et al. Expression of defective hepatitis B virus particles derived from singly spliced RNA is related to liver disease. J Infect Dis 2008;198:218-25. [Crossref] [PubMed]
- Kew MC. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J Gastroenterol Hepatol 2011;26:144-52. [Crossref] [PubMed]
- Chiu YT, Wong JK, Choi SW, et al. Novel pre-mRNA splicing of intronically integrated HBV generates oncogenic chimera in hepatocellular carcinoma. J Hepatol 2016;64:1256-64. [Crossref] [PubMed]
- Allain JP, Candotti D. Diagnostic algorithm for HBV safe transfusion. Blood Transfusion 2009;7:174-82. [PubMed]
- Warf MB, Berglund JA. The role of RNA structure in regulating pre-mRNA splicing. Trends Biochem Sci 2010;35:169-78. [Crossref] [PubMed]
- Huang FY, Wong DK, Seto WK, et al. Sequence variations of full-length hepatitis B virus genomes in Chinese patients with HBsAg-negative hepatitis B infection. PLoS One 2014;9:e99028 [Crossref] [PubMed]
- Chemin I, Alain S, Margeridon S, et al. What is really ongoing during occult HBV reactivation? Hepatology 2006;43:195-author reply 195-6. [Crossref] [PubMed]
Cite this article as: Candotti D, Allain JP. Biological and clinical significance of hepatitis B virus RNA splicing: an update. Ann Blood 2017;2:6.