
Limited potential of sheep as a large animal to model transfusion-related acute lung injury mediated by antibodies against human neutrophil antigen 3a
Introduction
Transfusion-related acute lung injury (TRALI) is a serious complication of blood transfusion. Although antibodies against several human neutrophil antigens (HNAs; including HNA-1, HNA-2 and HNA-3a) have been implicated in clinical cases of TRALI, antibodies against HNA-3a are frequently reported in severe cases (1-3). The HNA-3 antigen arises from a single nucleotide polymorphism in the SLC44A2 gene resulting in a single amino acid variation (c.461G>A, p.Arg154Gln) in choline transporter-like protein 2 (CTL2) (1). The high homology between human and mouse CTL2 protein (96%), which also expresses the Arg154 residue, suggested that human anti-HNA-3a antibodies could cross-react with mouse CTL2. This was demonstrated by Bayat et al. and Bougie et al. (4,5), with the former taking advantage of this cross-reactivity and inducing TRALI in mice with human anti-HNA-3a polyclonal antibodies isolated from donor sera.
Although small rodent models are crucial for scientific research, their clinical relevance may be limited due to differences in physiology, such as heightened respiratory rates and unique immunological characteristics. As a result, findings obtained from murine models may not be directly applicable to clinical settings (6). In addition, smaller size and lower blood volume of mice in comparison to humans restricts the amount of blood samples that can be collected, both in terms of volume and frequency. Currently, there are only four reports of in vivo TRALI models using larger animals: two studies each in pigs (7,8) and sheep (9,10). However, none of these models emulated TRALI mediated by anti-HNA-3a. Compared to mice, sheep exhibit greater similarity to humans in terms of anatomic, haemodynamic and microcirculatory characteristics, as well as erythrocyte life-span (11,12). Furthermore, our group has considerable experience with sheep biomedical models (13-15), including the TRALI models (9,10). Therefore, the development of an anti-HNA-3a mediated TRALI model using sheep could provide a useful tool in improving understanding of how TRALI develops. This study aimed to investigate whether human anti-HNA-3a alloantibodies bind to sheep CTL2, and thus whether sheep might be used to model anti-HNA-3a antibody mediated TRALI. Similarities in CTL2 between humans, mice and sheep were assessed using in silico analysis prior to evaluating whether human anti-HNA-3a alloantibodies bound to leucocytes from each species via flow cytometry. We present this article in accordance with the MDAR reporting checklist (available at https://aob.amegroups.com/article/view/10.21037/aob-23-57/rc).
Methods
In silico SLC44A2 gene and CTL2 protein comparison in humans, mice and sheep
Genetic comparison of SLC44A2 in humans, mice and sheep was performed using data from publicly available gene datasets on Ensembl genome browser (asia.ensembl.org/index.html). Corresponding protein analysis was performed using Universal Protein Resource (UniProt, https://www.uniprot.org/). Amino acid comparison was performed using protein sequences from UniProt with Clustal 2X (http://www.clustal.org/clustal2/). Clustal X (https://www.jalview.org/help/html/colourSchemes/clustal.html) was used to group and colour-code each amino acid residue based on their properties (e.g., hydrophobicity, positive charge, etc.). Protein sequence alignments were scored using the Gonnet PAM (point accepted mutation) 250 matrix (16).
Ethics, blood collection and sera preparation
This study was approved by the Australian Red Cross Lifeblood Ethics Committee (approval 2020#19) and the Queensland University of Technology UAEC (1500001013). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Lifeblood Ethics Committee of Australian Red Cross Lifeblood (No. EC00206). Informed consent was obtained from all individual participants. Experiments involving animals were performed under a project license (No. 1500001013) granted by the University Animal Ethics Committee (UAEC) of Queensland University of Technology, in compliance with Australian code for the care and use of animals for scientific purposes and the Animal Care and Protection Act. Genotyping of human volunteers for HNA-3 alleles was completed by Lifeblood’s Queensland Transplantation and Immunogenetics Services (QTIS) as outlined previously (17). Whole blood from humans, mice and sheep was collected as a source of leucocytes. After informed consent, 10–15 mL of whole blood was collected from healthy Lifeblood staff members (n=3) into ethylenediamine tetra-acetic acid (EDTA) vacutainer tubes (Becton Dickson, Franklin Lakes, NJ, USA). Trained animal facility staff collected whole blood from Merino sheep (10–15 mL; n=3) or from C57BL/6 mice (200 µL; n=3) into EDTA tubes. Sera known to contain anti-HNA-3a alloantibodies was also identified and provided by Lifeblood’s QTIS as outlined previously (17).
Leucocyte isolation and human lung microvascular endothelial cells (HLMVECs) preparation
Leucocytes were isolated from EDTA blood samples using IOTest 3 ammonium chloride-based erythrocyte lysing solution (Beckman Coulter, Brea, CA, USA) according to manufacturer’s instructions. HLMVECs (Cell Applications, Inc., San Diego, CA, USA) were cultured as previously described (17). Cultured HLMVECs were detached from culture flasks using 1.5 mL trypsin 0.05%-EDTA (Sigma Aldrich, St. Louis, MO, USA) before being resuspended in warm growth media (microvascular endothelial cell growth medium, Cell Applications, Inc.). Isolated leucocytes and detached HLMVECs were counted using a haemocytometer.
Binding of human anti-HNA-3a sera to human, mouse and sheep cells
Following leucocyte isolation or HLMVEC resuspension, 100,000 cells were resuspended with 25 µL of buffer [2% foetal bovine serum in phosphate buffered saline (PBS); Gibco, New York, NY, USA] before the addition of 25 µL anti-HNA-3a sera or negative sera [pool of sera from five donors which did not contain any anti-human leukocyte antigen (HLA) or HNA antibodies]. Cells were incubated for 30 min at 30 ℃ (±1 ℃) in dry incubator and centrifuged for 1 min at 150 g. Cells were washed by resuspended in 200 µL of buffer (2% foetal bovine sera in PBS; Gibco) and subsequently centrifuged for 5 min at 150 g. Following three washes, HLMVECs, sheep and mouse leucocytes were resuspended in 100 µL of buffer containing goat anti-human immunoglobulin G (IgG) (H+L) phycoerythrin (PE) conjugate (1:200 dilution; PA1-86078, Thermo Fisher, Waltham, MA, USA) while human leucocytes were resuspended in 100 µL of buffer containing goat anti-human IgG (H+L) Alexa Fluor 594 conjugate (1:100 dilution; A-11014, Thermo Fisher). Cells were stained in the dark for 30 min at room temperature before a further three washes with buffer. Cells were resuspended in 250 µL of buffer and analysed by flow cytometry (FACSAria, FACSDiva Software; Becton Dickson).
Results
A high homology (88% gene, 96% protein) between human and mouse SLC44A2 had been previously reported (4,18). In the present study we extended this by comparing publicly available SLC44A2 gene and CTL2 protein sequences for humans, mice and sheep (Table 1). Mouse and sheep SLC44A2 transcripts contained 16 exons, while human SLC44A2 transcripts contained 23 exons. Although several transcription variants were identified in both human and mouse SLC44A2, of these only three functional variants were recorded in humans, two in mice and one in sheep. Two matching variants in human and mouse SLC44A2 relate to an alternative upstream promoter that leads to an alternative exon 1 and a subsequent variation in the first 10–12 amino acids of CTL2 (4,19). These CTL2 isoforms are classified TV1 (706 amino acids) and TV2 (704 amino acids) (20,21). The third isoform (711 amino acids), identified in humans only, occurs due to splice variation and is identical to CTL2-TV2 in all but exon 16 (amino acids 672–706). Only one isoform was identified in sheep (705 amino acids). This isoform more closely resembled the TV1 (706 amino acid) variant of CTL2 in humans (i.e., 96% conservative protein similarity to human CTL2-TV1 vs. 95% conservative protein similarity to human CTL2-TV1). The longer TV1 variant is the only CTL2 isoform expressed on neutrophils, mononuclear cells, and platelets (4). Therefore, protein comparisons between the three species were performed using the TV1 variant (Table 1). Protein sequence analyses indicated a high similarity between human, mouse and sheep CTL2-TV1/CTL2. All three species shared a high protein homology both with (95–96%) and without (89–91%) conservative amino acid substitution predictions.
Table 1
| Characteristic | Human (Homo sapiens) | Sheep (Ovis aries) | Mouse (Mus musculus) |
|---|---|---|---|
| Gene location/size | |||
| Chromosome | 19 | 5 | 9 |
| Chromosomal location | 10,602,457–10,644,557 | 12,796,201–12,814,875 | 21,320,698–21,355,028 |
| Sense | Forward strand | Forward strand | Forward strand |
| Exons | 23 | 16 | 16 |
| Variation | |||
| No. of known splice variants [functional] | 15 [2] | 1 [1] | 10 [2] |
| Functional transcript IDs [protein length] | ENST0000335757.10 [706 aa]† | ENSOART00000018269.1 [705 aa]† | ENSMUST00000034697.7 [706 aa]† |
| ENST00000407327.8 [704 aa] | ENSMUST000000217461.1 [704 aa] | ||
| ENST00000586078.5 [711 aa] | |||
| Known PTV and missense summary‡,§ | |||
| Splice acceptor | 112 | 0 | 0 |
| Splice donor | 70 | 0 | 0 |
| Stop gained | 53 | 0 | 0 |
| Frameshift | 86 | 0 | 0 |
| Missense variant | 2,077 | 4 | 4 |
| Known synonymous variant summary‡,§ | 977 | 18 | 41 |
| Protein BLAST analysis (pairwise) | |||
| % identical match amino acid residues of human SLC44A2 to query protein [% conservative substitutions]¶ | – | 90 [95] | 91 [96] |
| % identical match amino acid residues of sheep SLC44A2 to query protein [% conservative substitutions]¶ | 90 [95] | – | 89 [95] |
| % identical match amino acid residues of mouse SLC44A2 to query protein [% conservative substitutions]¶ | 91 [96] | 89 [95] | – |
The table includes summary of protein variations (and transcript IDs) and pairwise comparison of CTL2 protein between the three species. Protein BLAST analysis performed using CTL2-P1 (406 aa variants of human and mouse CTL2; marked with “†”). ‡, transcript ID used as reference for variant summaries (marked with “†”) in functional transcript ID column; §, mutation summaries based on transcript information available on Ensembl (EMBL-EBI). Lack on mutation information may be a reflection on available genome data and not necessarily true reflection of genomic variation; ¶, UniprotKB IDs used for protein amino acid sequence. ID, identity; PTV, protein-truncating variant; CTL2, choline transporter-like protein 2.
Individual amino acid residue differences were assessed through protein sequence alignments between human, mouse and sheep CTL2-TV1 (Figure 1). Several other variations in cytosolic and transmembrane regions of CTL2-TV1 between the three species were found (Figure 1). In the first extracellular (EC) loop, only the Arg154 residue was identified in both mouse and sheep, suggesting that only the HNA-3a variant of CTL2 exists in these species.

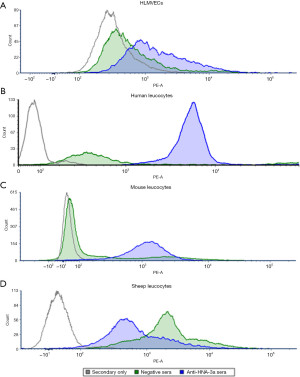
Cross-reactivity between human anti-HNA-3a antibodies and mouse CTL2 has previously been reported (4). This, in combination with the high homology between human, mouse and sheep SLC44A2/CTL2 sequences observed above, prompted flow cytometric investigation of the cross-reactivity between human anti-HNA-3a antibodies and sheep CTL2 compared to with human and mouse CTL2 (Figure 2). As expected, HLMVECs, human leucocytes, and mouse leucocytes all reacted with the anti-HNA-3a antibody containing sera but not with negative control sera (Figure 2A-2C, respectively). In contrast, sheep leucocytes reacted with the negative control sera to a greater extent than with the anti-HNA-3a containing sera (Figure 2D).
Discussion
In the present study, comparisons of SLC44A2 and CTL2 in humans, mice and sheep showed a high level of homology (88–89% gene homology, 95–96% protein homology). Individual amino acid differences in the EC loops of CTL2 showed that sheep domains were more homologous to human CTL2 than the sheep protein was to mouse CTL2. Only a single variant, that more closely resembled the longer TV2 variant, was observed in sheep. Furthermore, only an Arg154/HNA-3a variant was identified in sheep. This suggested that sheep CTL2 expressed on leucocytes might react with human anti-HNA-3a antibodies and therefore sheep may be a suitable animal in which to model anti-HNA-3a antibody mediated TRALI. This is important, because as previously suggested (22), replication of key findings in multiple model systems would strengthen their clinical relevance. Large animals, such as sheep, are more similar to humans than small animals such as mice (6) which have already been used to model anti-HNA-3a antibody mediated TRALI. However, as large animal models are expensive to develop, an important first step is to compare their SLC44A2 and CTL2 sequences with those in humans.
The next step then was to investigate cross-reactivity between sheep leucocytes and human anti-HNA-3a antibodies using sera previously shown to contain anti-HNA-3 antibodies (17). Two distinct classes of anti-HNA-3a antibodies, designated type I and type II, have been reported (23-25), with only the former being cross-reactive with CTL2 expressed on mouse leucocytes (5). This antibody type refers to the epitope of the antibody, with type I antibodies binding to the first EC loop CTL2, while type II antibodies require the expression of the first three EC loops on a cell membrane for proper epitope formation (5). In the present study, human and mouse leucocytes both reacted with the anti-HNA-3a containing sera, suggesting that it contained type I anti-HNA-3a antibodies. However, cross-reactivity between this sera and sheep leucocytes was not observed. The reactivity observed between the anti-HNA-3a antibody containing sera and sheep leucocytes was lower than that observed with control sera which did not contain any anti-HNA or anti-HLA antibodies. This could possibly be due to the sheep leucocytes reacting with non-specific IgG present in the control sera; however, no such non-specific binding was observed with either human or mouse cells. This ‘non-specific’ IgG is possibly xenoantibodies (such as anti-carbohydrate antibodies) which are present in all human sera from non-immunocompromised individuals (26). As xenoantibody titre varies from individual (and cognate antigen expression varies between species), it is possible that the control sera tested had a higher portion of xenoantibodies against sheep leucocytes than the anti-HNA-3a containing sera.
These results suggested that sheep are unlikely to be a suitable animal in which to model anti-HNA-3a mediated TRALI. It is possible that investigation of a wider panel of sera containing both type I and type II anti-HNA-3a antibodies might demonstrate cross-reactivity with sheep leucocytes. However, to be suitable for use in an in vivo model, such sera would need to be available in sufficient volumes to transfuse into multiple animals. In the absence of an anti-HNA-3a monoclonal antibody, specific isolation of anti-HNA-3a IgG from human sera (4) is another possible option; however, again large volumes of sera would be required. Based upon these results, the present study did not progress to complete functional in vitro experiments relevant to TRALI pathophysiology, nor were actual in vivo experiments with sheep considered at this stage. This limitation could be addressed by future experiments investigating if priming with an inflammatory stimulus such as lipopolysaccharide might precipitate anti-HNA-3a binding to sheep cells, as this has been shown to enhance binding of anti-HLA class I antibodies to human endothelial cells (27) and induce expression of HLA class II antigens on human neutrophils (28).
Another potential alternative large animal would be pigs as they have already been used to model TRALI mediated by other factors (7,8). Their suitability to model anti-HNA-3a antibody mediated TRALI could be explored in the future. Initial comparisons showed that pig CTL2 also shares a high homology to human CTL2 (92% non-conservative, 96% conservative homology). However, as we have demonstrated with sheep, laboratory experiments would be required to demonstrate cross-reactivity between pig CTL2 and human anti-HNA-3a antibodies. Interestingly, there are no reports of the Glu154 residue (HNA-3b variant) in pigs either, suggesting that this polymorphism is specific to humans.
Conclusions
In conclusion, investigation of sheep SLC44A2 and CTL2 sequences suggested the presence of Arg154/HNA-3a and showed strong homology compared to human SLC44A2/CTL2. However, leucocytes from sheep were not cross-reactive with human sera containing an anti-HNA-3a antibody. These results suggested that sheep would not be a suitable large animal in which to model TRALI mediated by human anti-HNA-3a antibodies.
Acknowledgments
The authors acknowledge Fenny Chong and Kelly Rooks of Lifeblood for the organization of human blood collections and phlebotomies. The authors also acknowledge both Emily Wilson from the Critical Care Research Group and Nadya Panagides of University of Queensland Biological Resources for their assistance with the blood collection from sheep and mice, respectively. The authors would also like to thank Eileen Roulis for her guidance regarding gene analysis.
Funding: This project utilised infrastructure provided by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://aob.amegroups.com/article/view/10.21037/aob-23-57/rc
Peer Review File: Available at https://aob.amegroups.com/article/view/10.21037/aob-23-57/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://aob.amegroups.com/article/view/10.21037/aob-23-57/coif). R.F. serves as an unpaid editorial board member of Annals of Blood from April 2017 to April 2026. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Australian Red Cross Lifeblood Ethics Committee (approval 2020#19) and the Queensland University of Technology UAEC (1500001013). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Lifeblood Ethics Committee of Australian Red Cross Lifeblood (No. EC00206). Informed consent was obtained from all individual participants. Experiments involving animals were performed under a project license (No. 1500001013) granted by the University Animal Ethics Committee (UAEC) of Queensland University of Technology, in compliance with Australian code for the care and use of animals for scientific purposes and the Animal Care and Protection Act.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Greinacher A, Wesche J, Hammer E, et al. Characterization of the human neutrophil alloantigen-3a. Nat Med 2010;16:45-8. [Crossref] [PubMed]
- Reil A, Keller-Stanislawski B, Günay S, et al. Specificities of leucocyte alloantibodies in transfusion-related acute lung injury and results of leucocyte antibody screening of blood donors. Vox Sang 2008;95:313-7. [Crossref] [PubMed]
- Davoren A, Curtis BR, Shulman IA, et al. TRALI due to granulocyte-agglutinating human neutrophil antigen-3a (5b) alloantibodies in donor plasma: a report of 2 fatalities. Transfusion 2003;43:641-5. [Crossref] [PubMed]
- Bayat B, Tjahjono Y, Sydykov A, et al. Anti-human neutrophil antigen-3a induced transfusion-related acute lung injury in mice by direct disturbance of lung endothelial cells. Arterioscler Thromb Vasc Biol 2013;33:2538-48. [Crossref] [PubMed]
- Bougie DW, Peterson JA, Kanack AJ, et al. Transfusion-related acute lung injury-associated HNA-3a antibodies recognize complex determinants on choline transporter-like protein 2. Transfusion 2014;54:3208-15. [Crossref] [PubMed]
- Simonova G, Tung JP, Fraser JF, et al. A comprehensive ovine model of blood transfusion. Vox Sang 2014;106:153-60. [Crossref] [PubMed]
- Patel NN, Lin H, Jones C, et al. Interactions of cardiopulmonary bypass and erythrocyte transfusion in the pathogenesis of pulmonary dysfunction in Swine. Anesthesiology 2013;119:365-78. [Crossref] [PubMed]
- Okazaki H, Ishikawa O, Iijima T, et al. Novel swine model of transfusion-related acute lung injury. Transfusion 2014;54:3097-107. [Crossref] [PubMed]
- Tung JP, Fraser JF, Nataatmadja M, et al. Age of blood and recipient factors determine the severity of transfusion-related acute lung injury (TRALI). Crit Care 2012;16:R19. [Crossref] [PubMed]
- Tung JP, Fung YL, Nataatmadja M, et al. A novel in vivo ovine model of transfusion-related acute lung injury (TRALI). Vox Sang 2011;100:219-30. [Crossref] [PubMed]
- Mock DM, Lankford GL, Burmeister LF, et al. Circulating red cell volume and red cell survival can be accurately determined in sheep using the [14C]cyanate label. Pediatr Res 1997;41:916-21. [Crossref] [PubMed]
- Brace RA, Langendörfer C, Song TB, et al. Red blood cell life span in the ovine fetus. Am J Physiol Regul Integr Comp Physiol 2000;279:R1196-204. [Crossref] [PubMed]
- Chemonges S, Shekar K, Tung JP, et al. Optimal management of the critically ill: anaesthesia, monitoring, data capture, and point-of-care technological practices in ovine models of critical care. Biomed Res Int 2014;2014:468309. [Crossref] [PubMed]
- Byrne L, Obonyo NG, Diab SD, et al. Unintended Consequences: Fluid Resuscitation Worsens Shock in an Ovine Model of Endotoxemia. Am J Respir Crit Care Med 2018;198:1043-54. [Crossref] [PubMed]
- See Hoe LE, Wildi K, Obonyo NG, et al. A clinically relevant sheep model of orthotopic heart transplantation 24 h after donor brainstem death. Intensive Care Med Exp 2021;9:60. [Crossref] [PubMed]
- Polyanovsky V, Lifanov A, Esipova N, et al. The ranging of amino acids substitution matrices of various types in accordance with the alignment accuracy criterion. BMC Bioinformatics 2020;21:294. [Crossref] [PubMed]
- Chiaretti S, Burton M, Hassel P, et al. Human neutrophil antigen 3 genotype impacts neutrophil-mediated endothelial cell cytotoxicity in a two-event model of TRALI. Blood Transfus 2022;20:465-74. [PubMed]
- Traiffort E, Ruat M, O'Regan S, et al. Molecular characterization of the family of choline transporter-like proteins and their splice variants. J Neurochem 2005;92:1116-25. [Crossref] [PubMed]
- Kommareddi PK, Nair TS, Thang LV, et al. Isoforms, expression, glycosylation, and tissue distribution of CTL2/SLC44A2. Protein J 2010;29:417-26. [Crossref] [PubMed]
- Kommareddi PK, Nair TS, Raphael Y, et al. Cochlin isoforms and their interaction with CTL2 (SLC44A2) in the inner ear. J Assoc Res Otolaryngol 2007;8:435-46. [Crossref] [PubMed]
- Flesch BK, Wesche J, Berthold T, et al. Expression of the CTL2 transcript variants in human peripheral blood cells and human tissues. Transfusion 2013;53:3217-23. [Crossref] [PubMed]
- Tung JP, Chiaretti S, Dean MM, et al. Transfusion-related acute lung injury (TRALI): Potential pathways of development, strategies for prevention and treatment, and future research directions. Blood Rev 2022;53:100926. [Crossref] [PubMed]
- Curtis BR, Sullivan MJ, Holyst MT, et al. HNA-3a-specific antibodies recognize choline transporter-like protein-2 peptides containing arginine, but not glutamine at Position 154. Transfusion 2011;51:2168-74. [Crossref] [PubMed]
- Berthold T, Wesche J, Kuhnert K, et al. Epitope mapping of antibodies directed against the human neutrophil alloantigen 3a. Transfusion 2011;51:2160-7. [Crossref] [PubMed]
- Woźniak MJ, Bowring C, Lucas G, et al. Detection of HNA-3a and -3b antibodies using transfected cell lines and recombinant proteins. Transfusion 2012;52:1458-67. [Crossref] [PubMed]
- Cooper DK. Xenoantigens and xenoantibodies. Xenotransplantation 1998;5:6-17. [Crossref] [PubMed]
- Morsing SKH, Zeeuw van der Laan E, van Stalborch AD, et al. A pulmonary endothelial amplification loop aggravates ex-vivo transfusion-related acute lung injury via increased toll-like receptor 4 and intra-cellular adhesion molecule-1 expression. Transfusion 2022;62:1961-6. [Crossref] [PubMed]
- Kelher MR, Banerjee A, Gamboni F, et al. Antibodies to major histocompatibility complex class II antigens directly prime neutrophils and cause acute lung injury in a two-event in vivo rat model. Transfusion 2016;56:3004-11. [Crossref] [PubMed]
Cite this article as: Radenkovic F, Burton M, Hassell P, Chiaretti S, Bui X, Zhao S, Jones M, Flower R, Tung JP. Limited potential of sheep as a large animal to model transfusion-related acute lung injury mediated by antibodies against human neutrophil antigen 3a. Ann Blood 2024;9:26.


