
RHD molecular analysis—from discovery to next generation sequencing
Introduction
Background
RHD genotyping is a critical part of the modern transfusion medicine diagnostic laboratory. D remains one of the most clinically significant blood group antigens in transfusion and in pregnancy and the significant number of D variants described to date compound many investigations requiring complex molecular analysis. This review focusses entirely on RHD but it should be appreciated that there is similar complexity surrounding RHCE variants. The review cites useful resources for cataloguing and describing the clinical significance of RHD variants, which genotyping methods must be able to define with a high degree of accuracy.
Rationale and knowledge gap
Several vulnerable patient groups are now supported significantly by RHD genotyping. In Europe, this has included the screening of many European pregnant D-negative women to genotype their fetuses in order to target the use of prophylactic anti-D to only pregnancies where D-positive infants are identified. The review describes these mass-scale applications of RHD genotyping which have been an outstanding success. Another vulnerable patient group which has been supported by RH (both RHD and RHCE) genotyping are multi-transfused patients, for example, in sickle cell disease. Where conventional genotyping technology is limited is that predominantly they require that the variant under investigation is previously known (mutations) so that polymerase chain reaction (PCR) amplification primers and probes can identify them. Next generation sequencing (NGS) has the potential to close this knowledge gap, it has no requirement to have previously defined mutations, with the exception that known RHD and RHCE targets are sequenced by gene-specific amplification. However, much is yet to be done to ensure that NGS and associated bioinformatics can support transfusion medicine laboratories effectively. These issues are discussed in this review.
Objective
We feel that the early pioneering years of RH molecular biology are often neglected in the literature when referring to the pathway leading to techniques that we all take for granted. We hope this review will address this and give due recognition to the early pioneering work that made this approach possible. Whilst the remainder of the review primarily focuses on NGS, brief mentions of other technologies that have emerged over the past twenty years (notably microarray platforms) are made. Finally, we have made reference to shortcomings in NGS as applied to RHD genotyping which lie predominantly in the bioinformatic based approaches to diagnosis. These are yet to be truly bespoke to RH and although the sequencing part of this approach is straightforward, the accurate reporting of RH variants has yet to be fully realized, especially considering that many novel variants still await discovery.
A brief history of Rh molecular analysis
The molecular era of Rh research emerged in the 1980s when Moore et al., and Carl Gahmberg almost simultaneously discovered that a 30–32 kDa red cell membrane protein was found in immunoprecipitates prepared from radio-iodinated red cells using anti-Rh sera (1,2). It was also realised five years later that these immunoprecipitates also contained a diffusely migrating (on SDS-PAGE gels) 45–50 kDa glycoprotein, later termed the Rh-associated glycoprotein, RhAG (3,4). In this relatively short time span, and somewhat serendipitously (as there was no direct proof at this stage), the major radio labelled 30–32 kDa component was assumed to carry all Rh antigens, and that RhAG was a tightly associated component that was invariably co-immunoprecipitated with anti-Rh. The emergence of monoclonal anti-D and other Rh specificities enabled two groups to purify both Rh proteins and RhAG to homogeneity sufficient to obtain N-terminal amino acid sequences (4,5). Using a different approach (hydroxyapatite chromatography), Peter Agre’s group of Johns Hopkins was also able to obtain N-terminal amino acid sequences of RhD proteins (6).
Following the determination of primary amino acid sequences of both Rh and RhAG in 1990 two groups described the isolation of cDNA clones of a RhCcEe mRNA (7,8). Two years later a RHD mRNA was cDNA cloned (9,10), and also that encoding RhAG (11). The publication of these sequences enabled the widespread assessment of Rh alleles, using reverse-transcriptase coupled PCR to isolate Rh cDNAs from peripheral blood drawn from individuals of known Rh phenotypes. Despite recent advances in NGS technologies (which this review will address), reverse transcription polymerase chain reaction (RT-PCR) followed by full length sequence analysis of Rh cDNAs still remains the gold standard method for the determination of Rh variants. In 1996 the molecular basis of the Rhnull phenotype was defined (12), point mutations in RHAG were demonstrated in such phenotype individuals showing the critical requirement of the RhAG in Rh complex assembly. Consequently, for a full assessment of a Rh variant, RHAG should be sequenced in addition to both RH genes. This is discussed later in this review.
RT-PCR based determination of Rh variants
At an early stage following Rh cDNA cloning, RT-PCR was used in combination with other molecular techniques to determine a number of RHD variants which had been described in the literature previously. Initially this included the clinically significant DVI variant (13) but rapidly included all of the D-categories DII, DIIIa, DIIIb, DIIIc, DIVa, DIVb, DVa, DVb, and DVII (14). In the early 1990s the original 7 D-category system was replaced by a nomenclature which was not consistent (three letter acronyms, e.g., DFR, DBT, DNU) which initially included a D followed by a descriptor which was either the initials of the country of origin, anti-D which defined the D variant or initials of the propositus (14-16). The International Society of Blood Transfusion (ISBT) has also published a complete list of RHD (004) alleles (www.ISBTweb.org/resource/004rhd.html).
Determination of RH gene structure
Shortly after the description of the first Rh cDNA, southern blotting using it as a probe determined that most D-negative individuals lacking the RHD gene are caused by a gross deletion (17). However, several D negative individuals were described that did not have this deletion. Gradually the molecular bases of these D-negative phenotypes emerged, including the common (in individuals of African ancestry) RHD pseudogene RHD*Ψ (18). Point mutations, hybrid RHD-RHCE genes, and small deletions were described and by the early 2000s it was realised that RH genetics was complex and that the various Rh phenotypes could readily be determined using molecular techniques (19). Workers also began to discover that the two genes RHD and RHCE had structural differences first described being an Alu repeat inserted in intron 4 of RHCE and that exon 10 of RHD was structurally larger (10,20). These physical differences opened up the availability of RHD genotyping assays which were utilized in the prenatal determination of fetal blood group in cases of RHD incompatibility leading to Hemolytic Disease of the Fetus and Newborn (HDFN). This eventually led to mass scale genotyping of whole European populations of RHD-negative women in order to preserve stocks of prophylactic anti-D where administration of it to mothers carrying D-negative fetuses is not required (21-24).
RH gene structure was further resolved when the ten intron/exon boundaries were defined (25). Then in 2000 Wagner et al., finally resolved the gross RH gene architecture based on progress in the human genome project and empirical experimentation (26). This paper described that RHD and RHCE were arranged in a tail to tail configuration and a gene TMEM50A (then termed SMP1) was located between them. TMEM50A appears to play a role in the expression of the products of the RHCE gene (27).
Weak and partial D
Following the cDNA cloning publications described earlier, research efforts were directed toward the molecular basis of weak D. Weak D has been operationally defined as a depression in D antigen site density but with no apparent loss of D epitopes, and thus no potential to generate anti-D following transfusion or pregnancy. This was in contrast to partial D, where D epitopes were missing, and clinically significant anti-D can be produced on exposure to normal D-positive red cells. Most partial D also have a marked depression in D antigen site density (28). Now we realise that this black/white definition does not have a precise boundary, as many examples of weak D have been described that do indeed produce anti-D (29). This has prompted several to suggest that the weak/partial D definition be abandoned and the term RH variant used instead (30). Early efforts to define the molecular basis of weak D had included a suggestion that a depression in RhD mRNA levels was responsible (31), later shown to be incorrect as described below.
In 1999 a breakthrough was achieved regarding the molecular basis of weak D. Using long-range PCR to amplify all ten RHD exons and flanking intronic regions and cDNA analysis Wagner et al. found that in all weak D samples investigated they possessed exonic mutations. This paper also described the molecular basis of weak D types 1–16 (32).
Since these pioneering studies describing the molecular background of partial and weak D phenotypes well over 150 weak D phenotypes, and over 100 partial D phenotypes have been described. This, along with multiple Del (D-elute) phenotypes (where D antigen expression is very weak indeed only detectable using sensitive adsorption-elution assays) shows a high degree of variability in Rh genetics. It is of clinical importance to establish the molecular backgrounds of these RH variants in order to inform both transfusion and pregnancy management in cases of alloimmunisation. By far the best resource available to those requiring information regarding RH variants is the Rhesus base website (http://www.rhesusbase.info) (33) which is curated by Dr. Franz Wagner.
The emergence of DNA microarrays
In 2005–2007 (34,35) two publications appeared describing the application of DNA microarrays and bead chip technology to the field of blood group genotyping (BGG). Both publications were supported by commercial manufacturers that brought products to the market [Progenika AG now part of Grifols (Derio, Spain) and Bioarray Solutions now part of Immucor (Warren, New Jersey, USA)]. Both platforms have subsequently been developed both involving probes to blood group specific single nucleotide variants (SNVs) attached to beads. Both of these platforms and others have found widespread application to inventory antigen-negative blood donations in blood banks and provide support to multi-transfused patients, notably sickle-cell disease (SCD) sufferers.
Array/bead chip technology clearly is of benefit to laboratories carrying out BGG. The technology is however limited to blood group-specific SNVs where the molecular basis has been previously described. This is an obvious limitation, but fortunately most clinically significant blood group antigens can be genotyped using them. For RH, it is clear that there are a significant number of variants still to be discovered, for example, we recently discovered 8 new variants in a Finnish D-negative pregnant female cohort (see later). Considering that RHD and RHCE are highly variable in individuals of African ancestry, where SCD is of major transfusion concern, higher resolution BGG of these genes is desirable. For this reason, direct sequence analysis using conventional NGS technology became desirable.
NGS based assessment of RH sequences
The long range-PCR approach adopted by Wagner et al., in 1999 (32) illustrated the relative simplicity of determining new RH variants by the analysis of sequences amplified directly from genomic DNA. In many cases the fresh samples normally required for RT-PCR analysis were unavailable. With the emergence of massively parallel sequencing or NGS technologies in 2005 (36), it was obvious that these techniques could be readily applied to the determination of RH variants. All NGS technologies require amplification of genomic targets, followed by their fragmentation [with the exception of single molecule sequencing (SMS), see later] and massively parallel sequencing of these fragments after their capture on beads. These sequences are then aligned to the human genome sequence, and then assembled utilising software into the final sequence readout. It is imperative that sufficient fragment populations are sequenced to cover the entire sequence, and read depth (numbers of times a particular base is sequenced) is a critical quality control in these experiments as misincorporations can then be identified and ignored, and that there are no gaps in the sequence reads. Single base mutations can then be readily identified by virtue of a roughly 50:50 split of the sequence read between two bases when compared to the human genome reference sequence.
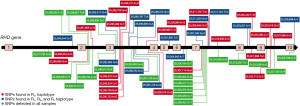
For NGS technology to function at all it is critical that the human genome sequence (or any target genome sequence for that matter) is available. Rather unfortunately the RHD reference sequence found in the hg19 and hg38 human genome builds (those most commonly used by NGS software) is a rare RHD variant DAU0 [ISBT RHD*10.00; encoded by c1136C.T (p.Thr379Met) in exon 8] which was soon realised (37). Thus, any analysis of normal RH genes when using the hg19 or hg38 builds will immediately identify DAU0-specific SNVs as apparent “mutations” when they are not. There are multiple DAU0-specific SNVs within its RHD gene (see Figure 1). It was therefore of critical importance to establish reference RHD and RHCE genes from individuals expressing the most common Rh phenotypes.
We and others cited here developed protocols for NGS of both RHD and RHCE and for the RHAG gene which has been shown to cause RH-ablation in Rhnull phenotypes. Several reports have involved the amplification of specific RHCE and RHD targets using allele-specific primers in order to amplify all or some RHD and RHCE exons and flanking intronic regions (38-44) whilst one paper reports using consensus RH primers (42). These studies have been directed to the management of patients with sickle cell disease but have, of course, wider implications to complex Rh analysis in patients and donors.
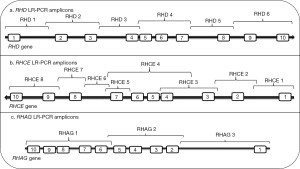
Our protocol is the only one described thus far in the literature to be capable of sequencing RHD in its entirety [out with whole genome sequencing (WGS)] and we briefly describe it here (44). Our method involves the amplification of the entire RHD gene in six overlapping long range-PCR fragments (Figure 2). These were then subjected to an Ion-Torrent PGM (personal gene machine) protocol (other platforms can of course be utilized) for NGS of these products. Initially, and as suggested above, we sequenced individuals expressing the most common RhD phenotypes to define reference sequences for RHD isolated from individuals expressing the most common Rh haplotypes Dce, DCe and DcE. In order to ensure that a single RHD gene was analysed these individuals were tested for RHD zygosity (45) using droplet digital PCR so that only hemizygous individuals were sequenced. We found that 38 SNVs, interspersed throughout the RHD gene, defined the differences between the Dce/DCe and DcE RHD genes. Interestingly we confirmed the earlier findings that the DAU0 gene corresponds to the hg19/38 human genome reference sequence, carries 21 intronic SNVs specific for DAU0 in addition to the exon 8 mutation encoding T379M which is common to all DAU phenotypes (46). Hg19/38/DAU0 also carries SNVs found in both RHD genes from DcE and DCe/Dce haplotypes (Figure 1). Thus, sequencing of any RHD variant with NGS technology and utilising hg19/38 as reference sequence will all need to consider the DAU0 gene mutations before identifying SNVs that would characterize any new variant.
We applied our NGS-RHD protocol to the analysis of 35 Finnish samples that had been identified by virtue of the fact they were serologically D-negative pregnant women but had detected the presence of fragments of RHD by conventional PCR-sequence specific primer (PCR-SSP) methods (47). NGS analysis revealed that 16 of these had previously been described as partial/D-negative/weak D/Del alleles. Fifteen of the remaining samples were found to encode 8 novel D variants. The remaining 4 samples failed to amplify various amplicon sets using our NGS-RHD method. In one of these samples an RHD amplicon described by (40), covering the region from exon 2 to exon 7 was amplified. It was found that part of intron 2 and the whole of exon 3 was deleted thus causing a D-negative phenotype. The other samples could not have their molecular basis determined due to the inability to amplify the entire RHD gene, but maybe have similar molecular backgrounds to the partial RHD gene deletion.
The phasing issue and SMS
The NGS protocol we adopted and that of the major commercial supplier of NGS technology, Illumina depends on short read sequencing of fragmented DNA products of the order of 200–400 bp. Whilst this method is readily applicable to samples obtained from hemizygous individuals there is an issue in defining maternally or paternally inherited mutations in homozygous RHD/RHD individuals. Due to the short reads of individual sequencing products, it is impossible to phase a number of mutations potentially spread across the entire RHD gene. SMS can address this issue with long >10 kb reads from the PCR amplicons used as starting material. We have used the MinION nanopore based sequencing platform to explore this issue (48). The results readily show the applicability to sequencing homozygous individuals but as 6 RHD-specific amplicons were used still does not completely address the phasing issue as mutations can occur on more than one amplicon. Zhang et al. (49) used a PacBio single molecule sequencer to investigate 11 individuals, 2 Caucasian and 9 with African ancestry. They achieved reads of 2.1–2.9 kb and used phasing markers to establish RH haplotypes. Interestingly in an individual carrying the RHD*Ψ, they found a 5.6 kb translocation of RHD into RHCE encompassing part of intron 8, exon 8 and part of intron 9 using this methodology. This would remain invisible to conventional NGS, illustrating the resolving power of long-read NGS.
Limitations of NGS when analysing RHD genes
As discussed earlier, many RHD variants possess hybrid RHD/RHCE genes, for example, all described DVI phenotypes. This presents a challenge when using current NGS technology for sequence analysis. This is largely due to the requirement to amplify long-range PCR amplicons prior to sequencing. In order to ensure these products are only derived from RHD, the forward and/or reverse PCR primers are allele specific. Thus, in hybrid genes some products may fail to amplify. An approach using combinations of RHD and RHCE specific primers may address this but must be bespoke for the variant under investigation—rarely an option for a busy reference laboratory. This is furthermore complicated by the potential occurrence of RHCE variants that harbour portions of the RHD gene. WGS coupled with suitable algorithms to define hybrid RH gene variants may also resolve these situations.
As already outlined, the universal adoption of the human genome sequence assemblies hg19/hg38 in software supplied with all NGS platforms is not ideal for BGG approaches. The unfortunate finding that the human genome sequence for a RHD gene is the rare DAU0 allele over complicates analysis. Our group has also found that a rare JK gene which is the reference hg19/38 gene does not resemble the common JK*A or JK*B alleles also complicates the analysis of JK genotypes (Altayar, Madgett & Avent, unpublished). For BGG exploiting NGS to be efficient for all blood group genes, bespoke databases with reference sequences of known blood group genotypes would be helpful. Such an approach has been described (50), but only includes data from the 1000 Genomes project. WGS, coupled with suitable analytical algorithms, has been shown to have sufficient resolving power to define RH genotypes and has been described recently (51), however full genome sequencing still is at a cost of around $1,000 per genome. It would be prudent however to perform such analysis on multi-transfused patients or even routine repeat blood donors.
Conclusions
This review has primarily focussed on historical aspects of RHD gene cloning and analysis that have led to the new gold standard approach of RHD genotyping by NGS. It is important to stress that much RHD genotyping is performed by conventional methods such as allele-specific PCR, often in combination with real-time quantitative PCR (for example, in prenatal testing used extensively in Europe). We have discussed microarray technology which is high throughput with several commercial examples being available. Another widespread technique in many areas of genotyping is the MLPA (multiplex ligation-dependent probe amplification) technique developed by MRC-Holland in the early 2000s which has been applied to RHD and RHCE genotyping (52). Using this technique has shown that hybrid RHCE-RHD genes lie in cis to the RHD*Ψ.
NGS undoubtedly provides a powerful tool in the armoury of red cell genotyping laboratories. It especially provides the ability to determine unknown RH variants which are not possible using microarray/bead technology that require the SNV combinations to be known in advance. There are shortcomings in the software analysis for BGG with respect to reference sequence utilisation but could readily be addressed by a commercial supplier of NGS kits for BGG. Long-read sequencing (single molecule) does partially resolve the ability to phase a particular allele but still is over complicated when analysing homozygous RHD/RHD as one will need to assume any detected variant is being analysed in tandem with a normal RHD gene, which is not always the case. Zygosity determination does also give a great deal of analytical power when coupled with NGS and should be routinely performed on any sample undergoing investigation as hybrid genes can be identified using this technology.
NGS still has a way to go before it can be applied routinely in BGG, foremost is the requirement to define reference sequences for the common blood group alleles and their incorporation into suitable analytical software. Nevertheless, the evolution of this technology in the past 16 years has been very significant and should be considered by specialist reference laboratories. The abundance of undefined RH variants also remains apparent, and NGS offers the best high throughput solution to this ongoing issue.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Emilia Sippert, Carine Prisco Arnoni and Maria Rios) for the series “Novel RH Alleles” published in Annals of Blood. The article has undergone external peer review.
Peer Review File: Available at https://aob.amegroups.com/article/view/10.21037/aob-22-41/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://aob.amegroups.com/article/view/10.21037/aob-22-41/coif). The series “Novel RH Alleles” was commissioned by the editorial office without any funding or sponsorship. NDA received funding for PhD studentships and consumables from Saudi cultural bureau and declares the advice given and paid to NDA for Non-invasive testing of blood groups, and payment to NDA for pending case. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Moore S, Woodrow CF, McClelland DB. Isolation of membrane components associated with human red cell antigens Rh(D), (c), (E) and Fy. Nature 1982;295:529-31. [Crossref] [PubMed]
- Gahmberg CG. Molecular identification of the human Rho (D) antigen. FEBS Lett 1982;140:93-7. [Crossref] [PubMed]
- Moore S, Green C. The identification of specific Rhesus-polypeptide-blood-group-ABH-active-glycoprotein complexes in the human red-cell membrane. Biochem J 1987;244:735-41. [Crossref] [PubMed]
- Avent ND, Ridgwell K, Mawby WJ, et al. Protein-sequence studies on Rh-related polypeptides suggest the presence of at least two groups of proteins which associate in the human red-cell membrane. Biochem J 1988;256:1043-6. [Crossref] [PubMed]
- Bloy C, Blanchard D, Dahr W, et al. Determination of the N-terminal sequence of human red cell Rh(D) polypeptide and demonstration that the Rh(D), (c), and (E) antigens are carried by distinct polypeptide chains. Blood 1988;72:661-6. [Crossref] [PubMed]
- Saboori AM, Smith BL, Agre P. Polymorphism in the Mr 32,000 Rh protein purified from Rh(D)-positive and -negative erythrocytes. Proc Natl Acad Sci U S A 1988;85:4042-5. [Crossref] [PubMed]
- Chérif-Zahar B, Bloy C, Le Van Kim C, et al. Molecular cloning and protein structure of a human blood group Rh polypeptide. Proc Natl Acad Sci U S A 1990;87:6243-7. [Crossref] [PubMed]
- Avent ND, Ridgwell K, Tanner MJ, et al. cDNA cloning of a 30 kDa erythrocyte membrane protein associated with Rh (Rhesus)-blood-group-antigen expression. Biochem J 1990;271:821-5. [Crossref] [PubMed]
- Le van Kim C, Mouro I, Chérif-Zahar B, et al. Molecular cloning and primary structure of the human blood group RhD polypeptide. Proc Natl Acad Sci U S A 1992;89:10925-9. [Crossref] [PubMed]
- Arce MA, Thompson ES, Wagner S, et al. Molecular cloning of RhD cDNA derived from a gene present in RhD-positive, but not RhD-negative individuals. Blood 1993;82:651-5. [Crossref] [PubMed]
- Ridgwell K, Spurr NK, Laguda B, et al. Isolation of cDNA clones for a 50 kDa glycoprotein of the human erythrocyte membrane associated with Rh (rhesus) blood-group antigen expression. Biochem J 1992;287:223-8. [Crossref] [PubMed]
- Cherif-Zahar B, Raynal V, Gane P, et al. Candidate gene acting as a suppressor of the RH locus in most cases of Rh-deficiency. Nat Genet 1996;12:168-73. [Crossref] [PubMed]
- Mouro I, Le Van Kim C, Rouillac C, et al. Rearrangements of the blood group RhD gene associated with the DVI category phenotype. Blood 1994;83:1129-35. [Crossref] [PubMed]
- Avent ND, Reid ME. The Rh blood group system: a review. Blood 2000;95:375-87. [Crossref] [PubMed]
- Daniels G. Variants of RhD--current testing and clinical consequences. Br J Haematol 2013;161:461-70. [Crossref] [PubMed]
- Reid ME, Lomas-Francis C, Olsson ML. The blood group antigen factsbook. Academic Press, 3rd edition, London and New York. ISBN: 9780124158498; 2012.
- Colin Y, Chérif-Zahar B, Le Van Kim C, et al. Genetic basis of the RhD-positive and RhD-negative blood group polymorphism as determined by Southern analysis. Blood 1991;78:2747-52. [Crossref] [PubMed]
- Singleton BK, Green CA, Avent ND, et al. The presence of an RHD pseudogene containing a 37 base pair duplication and a nonsense mutation in africans with the Rh D-negative blood group phenotype. Blood 2000;95:12-8. [Crossref] [PubMed]
- Wagner FF, Frohmajer A, Flegel WA. RHD positive haplotypes in D negative Europeans. BMC Genet 2001;2:10. [Crossref] [PubMed]
- Avent ND, Martin PG, Armstrong-Fisher SS, et al. Evidence of genetic diversity underlying Rh D-, weak D (Du), and partial D phenotypes as determined by multiplex polymerase chain reaction analysis of the RHD gene. Blood 1997;89:2568-77. [Crossref] [PubMed]
- Finning K, Martin P, Summers J, et al. Effect of high throughput RHD typing of fetal DNA in maternal plasma on use of anti-RhD immunoglobulin in RhD negative pregnant women: prospective feasibility study. BMJ 2008;336:816-8. [Crossref] [PubMed]
- de Haas M, Thurik FF, van der Ploeg CP, et al. Sensitivity of fetal RHD screening for safe guidance of targeted anti-D immunoglobulin prophylaxis: prospective cohort study of a nationwide programme in the Netherlands. BMJ 2016;355:i5789. [Crossref] [PubMed]
- Clausen FB, Steffensen R, Christiansen M, et al. Routine noninvasive prenatal screening for fetal RHD in plasma of RhD-negative pregnant women-2 years of screening experience from Denmark. Prenat Diagn 2014;34:1000-5. [Crossref] [PubMed]
- Haimila K, Sulin K, Kuosmanen M, et al. Targeted antenatal anti-D prophylaxis program for RhD-negative pregnant women - outcome of the first two years of a national program in Finland. Acta Obstet Gynecol Scand 2017;96:1228-33. [Crossref] [PubMed]
- Chérif-Zahar B, Le Van Kim C, Rouillac C, et al. Organization of the gene (RHCE) encoding the human blood group RhCcEe antigens and characterization of the promoter region. Genomics 1994;19:68-74. [Crossref] [PubMed]
- Wagner FF, Flegel WA. RHD gene deletion occurred in the Rhesus box. Blood 2000;95:3662-8. [Crossref] [PubMed]
- Chen H, Lv P, Liu Z, et al. Preliminary study on the function of TMEM50A and its correlation with the RH genes. Transfus Med 2021;31:277-85. [Crossref] [PubMed]
- Jones JW, Lloyd-Evans P, Kumpel BM. Quantitation of Rh D antigen sites on weak D and D variant red cells by flow cytometry. Vox Sang 1996;71:176-83. [Crossref] [PubMed]
- Sandler SG, Chen LN, Flegel WA. Serological weak D phenotypes: a review and guidance for interpreting the RhD blood type using the RHD genotype. Br J Haematol 2017;179:10-9. [Crossref] [PubMed]
- Daniels G, Poole G, Poole J. Partial D and weak D: can they be distinguished? Transfus Med 2007;17:145-6. [Crossref] [PubMed]
- Rouillac C, Gane P, Cartron J, et al. Molecular basis of the altered antigenic expression of RhD in weak D(Du) and RhC/e in RN phenotypes. Blood 1996;87:4853-61. [Crossref] [PubMed]
- Wagner FF, Gassner C, Müller TH, et al. Molecular basis of weak D phenotypes. Blood 1999;93:385-93. [Crossref] [PubMed]
- Wagner FF, Flegel WA. The Rhesus Site. Transfus Med Hemother 2014;41:357-63. [Crossref] [PubMed]
- Avent ND, Martinez A, Flegel WA, et al. The BloodGen project: toward mass-scale comprehensive genotyping of blood donors in the European Union and beyond. Transfusion 2007;47:40S-6S. [Crossref] [PubMed]
- Hashmi G, Shariff T, Seul M, et al. A flexible array format for large-scale, rapid blood group DNA typing. Transfusion 2005;45:680-8. [Crossref] [PubMed]
- Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005;437:376-80. [Crossref] [PubMed]
- Lane WJ, Westhoff CM, Gleadall NS, et al. Automated typing of red blood cell and platelet antigens: a whole-genome sequencing study. Lancet Haematol 2018;5:e241-e251. [Crossref] [PubMed]
- Fichou Y, Mariez M, Le Maréchal C, et al. The experience of extended blood group genotyping by next-generation sequencing (NGS): investigation of patients with sickle-cell disease. Vox Sang 2016;111:418-24. [Crossref] [PubMed]
- Dezan MR, Ribeiro IH, Oliveira VB, et al. RHD and RHCE genotyping by next-generation sequencing is an effective strategy to identify molecular variants within sickle cell disease patients. Blood Cells Mol Dis 2017;65:8-15. [Crossref] [PubMed]
- Schoeman EM, Roulis EV, Liew YW, et al. Targeted exome sequencing defines novel and rare variants in complex blood group serology cases for a red blood cell reference laboratory setting. Transfusion 2018;58:284-93. [Crossref] [PubMed]
- Jakobsen MA, Dellgren C, Sheppard C, et al. The use of next-generation sequencing for the determination of rare blood group genotypes. Transfus Med 2019;29:162-8. [Crossref] [PubMed]
- Stef M, Fennell K, Apraiz I, et al. RH genotyping by nonspecific quantitative next-generation sequencing. Transfusion 2020;60:2691-701. [Crossref] [PubMed]
- Lopez GH, McGowan EC, Condon JA, et al. Genotyping by sequencing defines independent novel RHD variants for an antenatal patient and a blood donor. Transfusion 2017;57:2281-3. [Crossref] [PubMed]
- Tounsi WA, Madgett TE, Avent ND. Complete RHD next-generation sequencing: establishment of reference RHD alleles. Blood Adv 2018;2:2713-23. Erratum in: Blood Adv 2019;3:120. [Crossref] [PubMed]
- Sillence KA, Halawani AJ, Tounsi WA, et al. Rapid RHD Zygosity Determination Using Digital PCR. Clin Chem 2017;63:1388-97. [Crossref] [PubMed]
- Wagner FF, Ladewig B, Angert KS, et al. The DAU allele cluster of the RHD gene. Blood 2002;100:306-11. [Crossref] [PubMed]
- Tammi SM, Tounsi WA, Sainio S, et al. Next-generation sequencing of 35 RHD variants in 16 253 serologically D- pregnant women in the Finnish population. Blood Adv 2020;4:4994-5001. [Crossref] [PubMed]
- Tounsi WA, Lenis VP, Tammi SM, et al. Rh Blood Group D Antigen Genotyping Using a Portable Nanopore-based Sequencing Device: Proof of Principle. Clin Chem 2022;68:1196-201. [Crossref] [PubMed]
- Zhang Z, An HH, Vege S, et al. Accurate long-read sequencing allows assembly of the duplicated RHD and RHCE genes harboring variants relevant to blood transfusion. Am J Hum Genet 2022;109:180-91. [Crossref] [PubMed]
- Möller M, Jöud M, Storry JR, et al. Erythrogene: a database for in-depth analysis of the extensive variation in 36 blood group systems in the 1000 Genomes Project. Blood Adv 2016;1:240-9. [Crossref] [PubMed]
- Chang TC, Haupfear KM, Yu J, et al. A novel algorithm comprehensively characterizes human RH genes using whole-genome sequencing data. Blood Adv 2020;4:4347-57. [Crossref] [PubMed]
- Haer-Wigman L, Veldhuisen B, Jonkers R, et al. RHD and RHCE variant and zygosity genotyping via multiplex ligation-dependent probe amplification. Transfusion 2013;53:1559-74. [Crossref] [PubMed]
Cite this article as: Madgett TE, Tounsi WA, Halawani AJ, Avent ND. RHD molecular analysis—from discovery to next generation sequencing. Ann Blood 2023;8:36.


