
Serology and molecular biology of DEL: a narrative review
Introduction: the importance of Del
The designation “Del” relates to an antigen D positive phenotype in which the presence of antigen D can only be demonstrated by adsorption and elution of anti-D while all conventional tests for antigen D including testing with anti-D in the indirect antiglobulin test (IAT) are negative (1) [in line with Association for the Advancement of Blood and Biotherapies (AABB) suggestions, in this review, Del is used for the phenotype and DEL for the allele].
The Del phenotype was first described by Okubo et al. (2) in 1984. They noticed that “some D negative red cells, though they were negative in a Du test after exposure to anti-D, could bind anti-D and yield it on elution” (2) and called these red blood cells (RBC) Del. Although their publication was officially only a “letter”, most of the properties nowadays associated with the most prevalent DEL allele in East Asia, RHD*01EL.01 have already been correctly identified: (I) in their (Japanese) population, about 10% of seemingly D-negative (D−) samples showed a Del phenotype; (II) the phenotype was most often associated with the presence of antigen C: About 53% of C positive but only 3% of C negative samples were Del; (III) no Del was detected in individuals who had formed an anti-D suggesting that Del individuals were unlikely to produce an anti-D.
Almost 40 years later, the interest in DEL remains current because red cell genotyping enables a precision medicine approach for patients and donors with DEL variants. Key issues concerning Del are:
- In East Asian populations, a relevant fraction of seemingly D− patients and blood donors possess a Del phenotype. For example, in a large Chinese population study (3), only 1,585 of 400,253 probands (0.4%) were D− in routine serology, and 275 (18%) of them displayed a Del phenotype. In these populations, D− RBC units are scarce and their availability is affected by the transfusion strategy for Del patients: is transfusion with D-positive (D+) RBC units safe for Del patients? What is the best approach to identify Del patients?
- Blood donors with a Del phenotype are typed as D− with routine methods like direct agglutination by anti-D or demonstration of antigen D in the IAT. Examples of anti-D immunization caused by Del units have been reported (4). Is it necessary to screen blood donors for Del to prevent anti-D immunization by Del RBC units?
- Mothers with some DEL alleles may become anti-D immunized. How can the DEL allele of the mother be determined? Will her DEL allele allow for anti-D immunization?
- Finally, in these days of whole genome or whole exome sequencing, increasingly molecular data must be analyzed without any knowledge of the serologic D phenotype. Which alleles express a Del phenotype? Is it possible to predict a Del phenotype with sufficient reliability based on molecular data alone if a previously unknown allele is detected in a proband?
Addressing these issues is crucial to define a rational transfusion strategy for patients and donors with Del phenotype. Numerous commentaries and reviews on the topic (5-11) have appeared. Recently, comprehensive reviews focusing on Del in China (10) and Japan (11) have been published. Details on specific alleles can be found in the Human RhesusBase (www.rhesusbase.info) (12) and in RHeference (www.rheference.org) (13). The International Society of Blood Transfusion (ISBT) nomenclature in this review is based on version 6.2 dated 30 September 2022 (https://www.isbtweb.org/resource/004rhd.html). As ISBT is constantly updating the tables, some alleles mentioned as “not listed” may have been included in more recent allele lists.
Here I give an overview on the serology and molecular basis of DEL with a specific focus on the remaining uncertainties in the structure-phenotype relationship and the differences between populations. This article was presented in accordance with the Narrative Review reporting checklist (available at https://aob.amegroups.com/article/view/10.21037/aob-22-16/rc).
Methods
A PubMed search with the terms “RHD, Del” was started (publication range 1984—the first description of the Del phenotype—to 2021; during revision a re-search was done until November 2022). “Related publications” search was done for publications related to the Del phenotype. The iterative approach was continued until no new relevant publications were identified. As second source, DEL alleles included in the Human RhesusBase (maintained by the author) and Rheference were analyzed for the references given. As third source, abstracts of the AABB and ISBT were screened for abstracts relevant for the Del phenotype using the term “RHD” to identify abstracts of possible relevance. For abstracts not matching previously identified publications, PubMed searches using abstract authors as key were done to identify possible related publications.
All identified publications in English defining new alleles or describing the prevalence of molecularly defined DEL alleles in a population were cited. Available publications giving information about immunization of Del patients or immunization by Del blood units were included if they were in English or German. Publications of mere technical merit (e.g., description of an assay for DEL) were not included if they did not add to the general understanding of the Del phenotype.
Non-English publications were considered if they were publicly available and contained critical information. If the same dataset was obviously published twice by the same authors in non-English and English journals, the publication in English was used as reference. If both a congress abstract and a peer-reviewed publication was identified, the peer-reviewed publication was cited.
The search strategy is summarized in Table 1.
Table 1
| Items | Specification |
|---|---|
| Date of search | 2021 (partial update 09-Nov-2022) |
| Databases and other sources searched | PubMed; Human RhesusBase; Rheference; ISBT RHD allele list; Abstracts of ISBT and AABB meetings |
| Search terms used | Del RHD (PubMed); RHD (abstracts) |
| Timeframe | 1984–2021 |
| Inclusion and exclusion criteria | All identified publications defining new DEL alleles, indicating a Del phenotype for a specific allele or describing the prevalence of molecularly defined DEL alleles in a population were included. Available publications giving information about immunization of Del patients or immunization by Del blood units were included if they were in English or German. Publications of mere technical merit (e.g., description of an assay for DEL) were not included if they did not add to the general understanding of the Del phenotype |
| Selection process | The selection process was performed by the single author |
ISBT, International Society of Blood Transfusion; AABB, Association for the Advancement of Blood and Biotherapies.
The Asian DEL
The high frequency of the Del phenotype among seemingly D− Japanese observed by Okubo et al. (2) was confirmed in other Asian populations: About 30% of seemingly D− donors (0.27%) in Hong Kong possessed a Del phenotype (14), resulting in a frequency of 0.079% among all donors. In contrast, in non-Asian populations, Del represents a minuscule fraction (0.26% or less) of seemingly D− blood donors (9).
There is now overwhelming evidence that the prevalent DEL allele in East Asian Del probands is RHD*01EL.01 carrying a “synonymous” c.1227G>A substitution (15-18). This allele was first dubbed RHD(K409K), later RHD(1227G>A) and is now often referred to as “Asia type” (19) or “Asian-type” (7) DEL. Curiously, this allele was first identified as cause of a Del phenotype in German blood donors (20). Its “synonymous” c.1227G>A substitution in codon 409 is immediately adjacent to the exon 9/ intron 9 junction and has a detrimental effect on RHD splicing: most RHD transcripts in RHD*01EL.01 lack RHD exon 9 (17,21,22).
Additional single nucleotide variations (SNVs) in intron 7 have been described for RHD*01EL.01-like DEL alleles: c.1073+152C>A (rs41307824) (15) and “c.1073+923C>T” (rs2427766) (21). The RHD*c.[1073+152C>A, 1227G>A] allele has been assigned a separate DEL allele number, RHD*01EL.36. A relevance of these intronic SNVs for the phenotype has been proposed (21) but never demonstrated. Most likely, these SNV do not impact the phenotype: c.1073+152C>A was detected by chance due to problems with a polymerase chain reaction (PCR) primer (15), c.1073+923T is the major allele at this SNV position among RHD alleles with a worldwide prevalence of 87% according to gnomAD (23).
RHD*01EL.01 is the most frequent DEL allele in East Asia (11,15-17,24-29) and in most populations with a relevant admixture of individuals of East Asian descent, like Australia (30) or the USA (31). In East Asia, up to a third of individuals who type D− by routine methods carry the RHD*01EL1.01 allele (32). Therefore, the question whether RHD*01EL.01 patients may safely be transfused with D+ units relevantly impacts blood supply for D− individuals in these countries. An overview of surveys of anti-D immunization in RHD*01EL.01 and other DEL alleles is given in Table 2 (19,33-35). No anti-D immunization was found among 358 pregnant women and 65 transfused patients with RHD*01EL.01. Two studies reported in parallel data on “true” RHD-negative patients: Combined, there were 99 anti-D among 483 pregnant women (20%) but none among 130 pregnant women with RHD*01EL.01 (P<0.001, Fisher’s exact test). Likewise, there were 11 anti-D among 160 RHD-negative transfused patients but none among 65 transfused patients with RHD*01EL.01 (P<0.05, Fisher’s exact test). In conclusion, there is no evidence of anti-D alloimmunization in RHD*01EL.01 patients in situations in which RHD-negative patients frequently develop an anti-D.
Table 2
| Population | D-negative (Del excluded) | RHD*01EL.01 | Other DEL alleles | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|
| Patients | Anti-D | Patients | Anti-D | Patients | Anti-D | ||||
| Pregnant women | 155 | 38 | 44 | 0 | (19) | ||||
| Pregnant women | 328 | 61 | 86 | 0 | 2 | 0 | (33) | ||
| Transfused patients | 160 | 11 | 65 | 0 | 2 | 0 | (33) | ||
| Pregnant women (Han Chinese) | 373 | No data | 130 | 0 | 12: RHD*01EL.44 (n=7); NL-8 (n=4); RHD*01N.07 (n=1) | 6: RHD*01EL.44 (n=3); NL-8 (n=2); RHD*01N.07 (n=1) | (34) | ||
| Pregnant women | 630 | No data | 98 (+70 patients excluded) | 0 | 10: RHD.01EL.02 (n=8); RHD*01EL.44 (n=1); NL-8 (n=1) | 2: RHD*01EL.44 (n=1); NL-8 (n=1) | (35) | ||
| Patients | 33 | 0 | Clinical trial NCT03727230* | ||||||
After several years of discussion among the experts (36), Shao (19) suggested a D+ transfusion strategy for Asian-type DEL based on the lack of documented anti-D immunization among RHD*01EL.01 probands and the absence of RHD*01EL.01 carriers among anti-D immunized individuals. The topic has recently been reviewed in much detail (10). D+ transfusion of patients with RHD*01EL.01 allows to set aside D− units for those patients who really need them (10,19,35,37). The safety of D+ transfusion for RHD*01EL.01 patients was further corroborated in a clinical trial of the Guangzhou Blood Center (NCT03727230, https://clinialtrials.gov/ct2/show/MCT0373730 accessed 26-May-2022): none of 33 Asian-type DEL recipients deliberately transfused with D+ blood developed an anti-D. It has even been suggested (38) that testing for RHD*01EL.01 should be extended to patients with rare weak D phenotypes, because an RHD*01EL.01 allele in trans would allow for D+ transfusion. While the logic of this policy is self-explanatory, the impact is limited: weak D and partial D probands are much rarer in China than individuals D− by routine serology [e.g., Yan et al. (39) observed in their donor cohort 1,401 D− donors but only 37 donors with weak D or partial D]. Furthermore, the likelihood of the presence of RHD*01EL.01 in weak D and partial D carriers is only about half of the likelihood in a seemingly D− individual.
The difficulties to discriminate the Del phenotype from other D phenotypes
Del or partial D?
In partial D, the RhD protein is changed in a way that D epitopes are lost and immunization to normal RhD seems possible. Typical mechanisms are RhD proteins in which segments are replaced by the corresponding RhCE segments, and RhD proteins with variations in the exofacial part of the protein (1). The “partial D” phenomenon is independent of the antigen density: partial D may occur with normal or even enhanced antigen density [e.g., RHD*03.04 (40)] as well as with reduced antigen density [e.g., RHD*06.02 (41,42)]. If a partial D antigen occurs in a Del phenotype, a “partial Del” phenotype results. While this concept seems intuitive at first glance, the verification that a Del phenotype expresses a partial D antigen may be painstakingly difficult and sometimes impossible:
Partial D phenotypes become obvious if an allo-anti-D is produced by its carrier. Such immunization events are rare, therefore often the lack of reactivity with monoclonal anti-D that cannot be explained by a low antigen density is used as indication of a partial D phenotype. Since a partial Del phenotype does not react with an anti-D in IAT, the investigation of D epitopes has to be accomplished by adsorption/elution tests using different monoclonal anti-D (43).
The first description of a partial Del phenotype dates from 2005, when Körmöczi et al. (43) obtained positive elution results for RHD*01EL.08 with 2 of 14 anti-D tested, contrasting with a uniform reactivity observed for RHD*01EL.01, RHD*01EL.35, and RHD*11, and an almost uniform reactivity in RHD*01EL.25. The identification of the absence of epitopes in partial DEL alleles is technically demanding. The absence or presence of reactivity with a specific antibody may depend on the method of detection used (44). It is symptomatic that RHD*11 was among the alleles without detectable epitope loss (43) while it is considered partial D by ISBT.
Based on serologic or molecular data, a partial Del status has been assumed in at least 14 of the DEL alleles listed by ISBT (RHD*01EL.04, RHD*01EL.05, RHD*01EL.08, RHD*01EL.09, RHD*01EL.19, RHD*01EL.22, RHD*01EL.23, RHD*01EL.31, RHD*01EL.33, RHD*01EL.42, RHD*01EL.44, RHD*01EL.47, RHD*01EL.48, RHD*01EL.49). Two of these alleles are also listed among the partial D (RHD01*01EL.22 as RHD*53 and RHD*01EL.23 as RHD*41).
Del or weak D?
At first glance, the discrimination of weak D and Del seems clear-cut: testing for antigen D in the indirect antiglobulin technique gives a positive result in weak D and a negative in Del. However, the result heavily depends on technical details precluding such simple distinction: Which technique has been used for the antiglobulin test? Which anti-D has been used for testing?
An antiglobulin test performed in tube technique is less sensitive than an antiglobulin test in gel (45) or even capture (46) technique. RHD*01W.49 was known as weak D but classified as Del when tested by IAT in tube technique (3). In the same study, RHD*01EL.06 and RHD*01EL.07 were serologically classified as Del, but the authors observed a stronger reactivity of the eluate than in other Del and reasoned that these alleles might qualify as weak D if tested in gel technique (3). In our laboratory, the fraction of RHD*11 samples that were missed by IAT diminished when we moved from tube technique to gel technique and on to solid phase technology.
The selection of the anti-D is critical, too. Anti-D differ in their avidity of binding to antigen D and in the D epitopes detected. In a partial Del phenotype, selection of the wrong anti-D will give a false negative result.
As additional layer of complexity, the antigen density of different samples carrying the same allele may vary. An RHCE*02 (Ce) allele in trans considerably reduces the antigen densities of weak D samples (40) and is likely to reduce the antigen density of Del samples. In addition, there is a yet unexplained “random variation” of the antigen densities of different samples with the same Rh phenotype (4,40). Even well characterized DEL alleles like the Asian-type DEL RHD*01EL.01 (32,47,48) and RHD*01EL.02 (48) are sometimes found to underlie a weak D phenotype.
Due to these confounders, the same allele may be classified as Del in one laboratory and as “weak” weak D in another. In fact, this happened for RHD*11 that was first described as weak D (49), then observed among Del samples (20) and later was listed among the partial D. The weak D alleles RHD*01W.58 (50) and RHD*01W.61 (3,47,51) have also been reported to encode a Del phenotype. In the case of RHD*01EL.46, the allele is listed by ISBT both among the DEL alleles as RHD*01EL.46 and among the weak D alleles as RHD*01W.94.
Antigen density of Del
A possibly more objective discrimination of Del and weak D might be based on the antigen density. Usually, antigen density is determined by flow cytometry (52). The measurement of D antigen density of Del samples is difficult, because there is a large overlap with the D-negative population. Weak D samples have antigen densities between 66 and 3,811 antigens/cell (40). Estimates for Del range from <22 to 22 in RHD*01EL.01 (43,46) and up to 50 antigens/cell in other Del-like types like NL-5 (50). Overall, the data are scarce and only a few Del phenotypes have been investigated (43,46,50,53) (see Table 3).
Table 3
| Allele | Antigen density | Reference |
|---|---|---|
| RHD*01EL.01 | <22 | (43) |
| RHD*01EL.01 | 22 | (46) |
| RHD*01EL.08 | <22 | (43) |
| RHD*01EL.11 | <22 | (43) |
| RHD*01EL.33 | 24 to 28 (median 26) | (53) |
| RHD*01EL.35 | <22 to 26 (median <22) | (43) |
| RHD*11 | <22 to 36 (median 33.5) | (43) |
| NL-5 (characterization by exon PCR) | 50 | (50) |
PCR, polymerase chain reaction.
Del or D-negative?
While partial D and Del are non-exclusive definitions and the overlap between weak D and Del is explained by the variable sensitivity of antigen D testing in indirect antiglobulin technique, the difficulties to discriminate Del from D− are of technical nature:
In a partial Del, use of a monoclonal anti-D directed to D epitopes absent in the partial Del type will result in a negative adsorption/elution test and the Del status may be missed if only this anti-D is used. As often only a minority of anti-D is binding, several Del have initially been characterized as D− (e.g., RHD*01EL.18, RHD*01N.07, RHD*01N.77) and their Del status revealed in later studies (11,34,54,55). Likewise, old data (56) on a high frequency of samples with RHD intron 4 and exon 10 in Japanese D− tested negative by adsorption/elution are difficult to reconcile with current knowledge if not assuming a limited sensitivity of the adsorption/elution tests performed.
On the other hand, use of low-ionic washing buffers in the elution process may lead to false positive results due to non-specific adsorption of high-titer antibodies (57). If residual donor samples are used for characterization, contamination might also be an issue: Many blood grouping machines like the Beckman-Coulter PK series (e.g., PK 7300) machines do not use single use tips for pipetting. D+ RBCs may carry as much as 40,000 antigens per cell while Del RBCs have 30 or less. A contamination of D− RBC by 0.1% D+ would result in a positive adsorption/elution test. In conclusion, false-positive adsorption/elution is a non-trivial problem: In a recent study in Korea (58), 5.5% of RHD deletion samples showed a positive adsorption/elution result. These technical considerations explain why the DEL status of several alleles is doubtful.
Regrettably, the molecular structure is not very helpful: As detailed below, missense variations may alternatively lead to weak D, Del or D− phenotypes, the impact of splice site variations may vary, and even variations expected to abolish normal RhD expression like frameshift variations (54) or deletions of whole exons (59) may result in a Del phenotype.
Single molecule fluorescence microscopy complemented by machine learning has been suggested as alternative to adsorption/elution for the discrimination of Del from D− (44). This approach is based on fluorescence microscopy and uses the information gained from the distribution of signals on the RBC’s surface to discriminate specific and non-specific binding. The suitability of the method to discriminate Del from D− was demonstrated with RHD*01EL.08 and RHD*09.05 samples which were also used for training of the algorithm. The method did not yet gain widespread use, possible due to the demanding technical requirements. It will be interesting to see whether it is less error-prone in routine use than adsorption/elution.
Molecular bases of Del
The first description of a correct molecular basis of a DEL allele was the result of a study in Germany (20) on donors harboring parts of the RHD gene but typed D− by direct agglutination and in IAT.
Seventeen different alleles were detected, three of which were demonstrated to encode a Del phenotype: RHD(M295I) (now known as RHD*11), RHD(K409K) with a c.1227G>A variation (nowadays RHD*01EL.01) and RHD(IVS3+1G>A), now known as RHD*01EL.08. Hence, missense variations and splice site variations were recognized as major molecular mechanisms leading to Del phenotypes already in this study.
Including the “Asian-type” DEL allele RHD*01EL.01, the current ISBT RHD allele table version 6.2 dated 30 September2022 lists 48 DEL alleles (Table 4) (3,4,15,20,27,30,51,53-55,59,63,64,66-69,71,76,79,81,83-85,87). In three of these alleles (RHD*01EL.32, RHD*01EL.35, RHD*01EL.37), the polymorphism indicated in the ISBT table as difference to normal RHD is unlikely to cause the Del phenotype, because the variations are frequent or even present (RHD*01EL.37) in the NCBI reference sequence NG_007494.1.
Table 4
| ISBT name | Nucleotide variations$ | Predicted protein variations$ | Haplotype/phenotypes$ | Mechanism | Comments | Reference |
|---|---|---|---|---|---|---|
| RHD*01EL.01 | c.1227G>A | p.= | DCe | Splice site variation (exon 9/IVS 9)‡ | Asian-type DEL, prevalent in East Asia | (20) |
| RHD*01EL.02 | c.3G>A | p.Met1? | Cce and cEe (3) | Loss of start codon | (51) | |
| RHD*01EL.03 | c.53T>C | p.Leu18Pro | Cce (3) | Single missense variation | (51) | |
| RHD*01EL.04 | c.[147del, 148+6del] | DCe | Deletion of A at position 147 causing frameshift | AA>A; the deletion of c.148+6A is not mentioned by ISBT but seemingly always present when investigated (54). Anti-D immunization reported (60) | (54) | |
| RHD*01EL.05 | c.148+1G>A | DcE | Splice site variation (exon 1/IVS 1)§§ | Mentioned in abstract form in 2001 (61)† | (30) | |
| RHD*01EL.06 | c.251T>C | p.Leu84Pro | cEe (3) | Single missense variation | Antigen density higher than Asian-type DEL, weak D excluded in tube technique only (3) | (51) |
| RHD*01EL.07 | c.410C>A | p.Ala137Glu | Cce (3) | Single missense variation | Antigen density higher than Asian-type DEL, weak D excluded in tube technique only (3) | (51) |
| RHD*01EL.08 | c.486+1G>A | DCe | Splice site variation (exon 3/IVS 3)§ | Partial Del phenotype (43). 19 amino acid insertion after Asn162 predicted (62) | (20) | |
| RHD*01EL.09 | c. 486+2T>A | DcE and DCe | Splice site variation (exon 3/IVS 3)§ | Del phenotype observed in CcDee (30) | (54) | |
| RHD*01EL.10 | c.1222T>C | p.Trp408Arg | DCe | Single missense variation | (27) | |
| RHD*01EL.11 | c. 1252dup | p.Ter418LeuextTer72 | DCe | Duplication of c.1252T with loss of stop codon | TTTT>TTTTT | (63) |
| RHD*01EL.12 | c.458T>C | p.Leu153Pro | cEe | Single missense variation | (54) | |
| RHD*01EL.13 | c.786del | DCe | Deletion of c.786A causing frameshift | AA>A; reported as D− (54) | (54) | |
| RHD*01EL.14 | c.634+5G>T | DCe | Splice site variation (exon 4/IVS 4)‡ | Reported as weak D (64) | (64) | |
| RHD*01EL.15 RHD*01N.52 |
c.922G>T | p.Gly308Ter | DCe | Nonsense variation | Reported as Del in abstract form (65)† probably due to false-positive adsorption/elution; most likely D− (66) | (66) |
| RHD*01EL.16 | c.634G>C | p.Gly212Arg | Dce | Missense (splice site exon 4/IVS 4 affected)‡ | (54) | |
| RHD*01EL.17; RHD*01N.22 | c.1203T>A | p.Tyr401Ter | DcE | Nonsense variation | Initially described as D− by routine serology. Del phenotype first reported by Flegel et al. (54) | (63) |
| RHD*01EL.18; RHD*01N.50 | c.93dup | p.Thr32TyrfsTer4 | DCe | Duplication of c.93T causing frameshift | TTTTTTT>TTTTTTTT; Del phenotype first reported by Flegel et al. (54) | (67) |
| RHD*01EL.19 | c.635-2A>G | Not reported | Splice site variation (IVS 4/exon 5)§ | Del phenotype mentioned in HE577129 | (68) | |
| RHD*01EL.20 | c.1154-8T>A | Not reported | Splice site variation (IVS 8/exon 9)‡ | No detailed serology available | (69) | |
| RHD*01EL.21 | c.148+5G>C | Cce | Splice site variation (exon 1/IVS 1)# | (30) | ||
| RHD*01EL.22; RHD*53 | c.336-2del | Cce | Deletion of 336-2A causing splice site variation (exon 2/IVS 2)§ | Partial Del phenotype according to genbank entry KC341996 | (30) | |
| RHD*01EL.23; RHD*DBU; RHD*41 | RHD-cE(5-7)-D | RhD-cE(5-7)-D | DcE | hybrid allele | Substitution of RHD exons 5 to 7 by the corresponding exon 5 to 7 of the cE allele of RHCE; partial Del phenotype expected | (54) |
| RHD*01EL.24 | c.838G>A | p.Ala280Thr | Cce | Single missense variation | No impact on exon 6 splicing in minigene splicing assay (70) | (55) |
| RHD*01EL.25 | c.1252T>A | p.Ter418LysextTer26 | DCe | Loss of stop codon | (26) | |
| RHD*01EL.26 | c.1247dup | p.Phe417IlefsTer73 | DCe | Duplication of 1247G causing frameshift | The allele is designated RHD*1248G in two manuscripts (71,72). The accompanying genbank entry KJ145906.1 displays an insertion before position 1248 (c.1247_1248 insG or c.1247dup). However, the ISBT table lists an insertion after position 1248 (c.1248_1249insG) which would result in p.Phe417ValfsTer73 | (71) |
| (RHD*01EL.27) | Not used* | |||||
| RHD*01EL.28 | c.993del | p.Phe332SerfsTer28 | not reported | Deletion c.993C causing frameshift | No phenotype reported in LN680540 | † |
| RHD*01EL.29 | c.1210G>C | p.Asp404His | DcE | Single missense variation | Reported as Del in JX114749 | † |
| RHD*01EL.30 | c.1074-649_1153+266del | p.359_384delins56$ | Not reported | Large deletion encompassing exon 8 | In the longest transcript, exon 8 (80 nucleotides) is replaced by an intron 7 segment comprising 170 nucleotides | (59) |
| RHD*01EL.31 | c.148+1G>T | IVS1+1G>T | DcE | Splice site variation (exon 1/IVS 1)‡‡ | Partial Del (73) | (71) |
| RHD*01EL.32 | c.149-29G>C | DCe | Intron polymorphism | This polymorphism is frequent in D+ alleles and not causative of the phenotype (74). According to gnomAD (23), the frequency of c.149-29G>C is 0.600. c.149-29G>C is typical for RHD alleles in Dce, DCe or DCE haplotypes (75) | (76) | |
| RHD*01EL.33 | c.336-2A>G | DCe | Splice site variation (IVS 2/exon 3)§ | (53) | ||
| (RHD*01EL.34) | Not used* | |||||
| RHD*01EL.35 | c.802-41_802-38del; c.802-38_35del (ISBT) | DCe | Intron polymorphism | An allele with a c.802-41_802-38 del was described in 2005 (4) and listed by ISBT as RHD*01EL.35 for many years. The polymorphism in IVS5 is frequent (77) and may not be the cause of the Del phenotype. According to gnomAD (23), the frequency of c.802-41_802-38del is 0.071. Since version 6.0 of the RHD allele tables, a different position of the deletion is indicated but no source given | (4) | |
| RHD*01EL.36 | c.[1073+152C>A,1227G>A] | DCe | Splice site variation (exon 9/IVS 9) | The polymorphism in IVS7 was detected because it interfered with primer binding; an influence on the phenotype is not documented | (15) | |
| RHD*01EL.37 | c.1154-31C>T | Not reported | Intron polymorphism | Mentioned in abstract form (65). The polymorphism in IVS8 is frequent and may not cause the phenotype. The T is present in the NCBI reference sequence NG_007494.1. According to gnomAD (23), the frequency of c.1154-31T is 0.775. c.1154-31C is typical for RHD alleles in a DcE haplotype (75) | † | |
| RHD*01EL.38; RHD*01N.57 | c.1010T>G | p.Leu337Arg | DCe | Single missense variation | Adsorption/elution not tested in original publication. Slightly reduced exon inclusion of exon 6 (80%) in minigene splicing assay (70) | (71) |
| RHD*01EL.39 | c.113T>A | p.Leu38Ter | Not reported | Nonsense variation | Reported as Del in abstract form (65)† probably due to false-positive adsorption/elution (78)† | † |
| RHD*01EL.40 | c.278T>G | p.Leu93Arg | Cce | Single missense variation | (71) | |
| RHD*01EL.41 | c.872C>G | p.Pro291Arg | Cce | Single missense variation | Slightly reduced exon inclusion of exon 6 (80%) in minigene splicing assay (70) | (79) |
| RHD*01EL.42 | c.[149-29G>C,335G>C] | p.Ser112Thr | Not reported | Missense (splice site affected exon 2/IVS 2)§ | (79) | |
| RHD*01EL.43 | c.46T>C | p.Trp16Arg | DcE | Single missense variation | Also reported as weakly positive in IAT (79) | (79,80) |
| RHD*01EL.44 | RHD-RHCE(4-9)-RHD | RhD-RhCE(4-9)-RhD | DCe | Hybrid allele | Partial Del (34) | (3) |
| RHD*01EL.45 | c.721A>C | p.Thr241Pro | Not reported | Single missense variation | (66) | |
| RHD*01EL.46 RHD*01W.94 |
c.884T>C | p.Met295Thr | DCe or DcE | Single missense variation | Initially described as weak D (81,82); later reported as Del (66) | (81) |
| RHD*01EL.47 | c.510dup | p.His171AlafsTer28 | cEe | Frameshift (duplication of 510G) | Partial Del (83) | (83) |
| RHD*01EL.48 | c.1154-412_1227+526del | Not reported | Large deletion encompassing exon 9 | Also designated DKG; partial Del (weakly positive with 4 of 12 anti-D tested) | (84) | |
| RHD*01EL.49 | c.1016G>T | p.Gly339Val | Cce | Single missense variation | Probably partial Del (85); loss of epD6.6, 8.2 and 9.1 | (85) |
| RHD*01EL.50 | c.1151C>G | p.Thr384Arg | Cce | Missense variation | First observed with “unknown phenotype” (86); Del phenotype shown in (87) | (87) |
| RHD*09.05 | c.[602C>G,667T>G,819G>A,872C>G] | p.[Thr201R,Phe223Val,Pro291Arg] | Dce | Multiple missense variations | Known as weak D type 4.3; structurally closely related to weak D type 4.0 (RHD*09.03.01). Phenotype is Del, no data on partial D phenotype | (88) |
| RHD*11 | c.885G>T | p.MetM295Ile | DCe | Single missense variation | Initially reported as weak D in a Dce haplotype (49). More frequent in DCe, usually with a borderline weak D/Del phenotype; allo-anti-D described. Slightly reduced exon inclusion of exon 6 (70%) in minigene splicing assay (70) | (20) |
| RHD*01N.07 | RHD-RHCE(4-7)-RHD | RhD-RhCE(4-7)-RhD | Cce | Hybrid allele | Initially reported as D− in DcE haplotype (89); later found in Chinese C+c+E-e+ Del sample (55). Partial Del (34) | (89) |
| RHD*01N.48 | c.822delG | p.Leu275TrpfsTer13 | Cce | Frameshift | Described as D− in genbank entry HG779212 but listed as adsorption/elution positive in the accompanying manuscript (79). Deletion is at position c.822, not c.882 | (79) |
| RHD*01N.60 | c.1213C >T | p.Gln405Ter | CcEe | Nonsense variation exon 9 | Eluate weakly positive (11) | (11) |
| RHD*01N.67 | c.1-15144_148+3158del | Dce | Large deletion exon 1 | Deletion characterized in Del sample (90) | (90) | |
| RHD*01N.77 | c.1228-1G>A | Cce | Splice site variation (IVS 9/exon 10)§§ | Eluate 2+ (11) | (71) | |
| RHD*01W.58 | c.1006G>C | p.Gly336Arg | Cce | Single missense variation | Very weak D, may appear as Del (50). Almost normal exon inclusion of exon 6 (>80%) in minigene splicing assay (70) | (69) |
| RHD*01W.61 | c.28C>T | p.Arg10Trp | DCe | Single missense variation | Generally known as weak D [AM412754, (82,91)]; reported as Del in China (3,47,51) | (51) |
| Not listed; NL-1 | c.1154-374_1227+563del | Not reported | Large deletion encompassing exon 9 | This 1,013 bp deletion is different from the deletion in RHD*01EL.58 and was initially described to underlie most Asian Del samples (92). If the allele exists it must be rare. | (92) | |
| Not listed; NL-2 | c.486+5G>A | cEe | Splice site variation (exon 3/IVS 3)§ | No serology in initial publication (69); characterized both as Del (79) and as weak D (93) | (69) | |
| Not listed; NL-3 | RHCE(1-8)-D(9-10) | DCe | Structure not fully characterized | RHD exons 1 to 7 are present but transcript analysis showed truncated RHD(1-7); RHCE(1-8)-RHD(9-10) and normal RHCE transcripts (94) | (94) | |
| Not listed; NL-4 | c.[602C>G, 667T>G, 819G>A, 919G>A] | p.[Thr201Arg,Phe223Val,Gly307Arg] | Not reported | Multiple missense variations | Characterized as Del in (66). Reduced exon inclusion of exon 6 (50%) in minigene splicing assay (70) | (95) |
| Not listed; NL-5 | c.1227+2874_1254+1317del | DCe | Large deletion exon 10 (RHD ex10 del type 1) | Initially described as weak D (96). Also observed in Del samples (79,93,97). The exact position of the deletion is not described in all reports relating to Del phenotype (79,93) | (96) | |
| Not listed; NL-6 | RHD(1-9)-RHCE(10) | p.= | DCe | Hybrid allele | Hybrid structure supported by cDNA analysis (76). As the protein structure is identical to RhD, the cause of the Del phenotype is unknown | (76) |
| Not listed; NL-7 | c.93T>A | p.Phe31Leu | Cce | Single missense variation | (55) | |
| Not listed; NL-8 | RHD-RHCE(2-5)-RHD | RhD-RhCE(2-5)-RhD | Not reported | Hybrid allele | Differs from DVI type IV (98) by a definitive RHCE origin of exon 2. Partial Del (34) | (55) |
| Not listed; NL-9 | c.487-1G>A | Cce | Splice site variation (IVS 4/exon 4) | (86) | ||
| Not listed; NL-10 | c.1027del | p.Tyr343ThrfsTer16 | cEe | Frameshift | (99) |
$The variations are described according to the HGVS recommendations for sequence variant nomenclature (100). The haplotypes (Dce, DcE, DCe or DCE) are indicated as reported in the reference or deduced from independent reports. If only a single sample or discordant samples were reported, the antigens C, c, E and e present in the samples are indicated [e.g., cEe if the sample was C-c+E+e+]. *RHD*01EL.27 and RHD*01EL.34 have never been used in an official ISBT DEL allele listing. #No impact on splicing in minigene splicing assay (101). ‡,‡‡Full length exon inclusion maintained in minigene splicing assay; references: ‡(102), ‡‡(101). §,§§No full length exon inclusion transcripts observed in mini-gene splicing assay (probable partial Del); references §(102), §§(101). †Described in abstract form or as genbank entry only. ISBT, International Society of Blood Transfusion; IVS, intervening sequence (Intron); IAT, indirect antiglobulin test.
Ten additional RHD alleles have been mentioned to be associated with a Del phenotype but are not acknowledged by ISBT as DEL and listed as partial D (RHD*09.05, RHD*11), weak D (RHD*01W.58, RHD*01W.61) or D− (RHD*01N.07, RHD*01N.48, RHD*01N.60, RHD*01N.67, RHD*01N77). The other way around, in five DEL alleles acknowledged by ISBT (RHD*01EL.13, RHD*01EL.15, RHD*01EL.20, RHD*01EL.28, RHD*01EL.39) the evidence for a Del phenotype is weak, because peer-reviewed publications or independent descriptions in abstract form are lacking. Finally, ten structures (NL-1 to NL-10) have been reported to be associated with a Del phenotype but have not been included in the ISBT allele tables yet.
About two thirds of DEL alleles harbor missense variations in RHD (15 DEL alleles listed by ISBT plus 6 other) or splice site variations (14 listed by ISBT plus 3 other). The remaining third (18 listed by ISBT plus 10 other) are caused by many different mechanisms: Rearrangements of the RH locus including RHD/RHCE hybrid alleles (2 listed by ISBT plus 4 other), large deletions encompassing at least one RHD exon, variations in the termination codon leading to an elongated protein, variations in the start codon, duplications or small deletions introducing a frameshift and nonsense variations introducing a premature stop codon. The listing of molecular causes is certainly incomplete: the alterations described in three alleles cannot be the cause of a Del phenotype and there have been repeated observations (26,54,66,99) of a Del phenotype in samples with a seemingly normal RHD gene indicating that some causes of DEL cannot be found by currently used methods.
DEL alleles with missense variations
There are 21 alleles expressing a Del phenotype that carry a single missense variation. Two additional alleles carry a missense variation on a weak D type 4.0 (RHD*09.03.01) background. Similar to weak D alleles (49), the missense variations are located in the intracellular or transmembrane segments of the RhD protein [Figure 1 (103)].
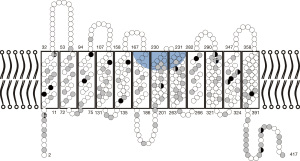
Missense variations may lead to a reduced antigen density by two non-exclusive mechanisms (listed in the order of interference with protein synthesis):
(I) Sometimes, the altered nucleotide sequence may interfere with correct splicing. In two of the 23 alleles (RHD*01EL.16 and RHD*01EL.42), the missense variation is adjacent to an exon/intron junction and the Del phenotype most likely caused by an effect on splicing (102). However, even SNV in the exon not directly adjacent to an intron/exon junction may cause incorrect splicing (70,104).
(II) Missense variations impact the protein structure and may disrupt protein folding, intramolecular interactions, and intermolecular interactions with partners leading to reduced protein integration into the membrane or antigen loss. When the effect of the 23 Del-associated missense variations on protein structure is evaluated using prediction tools like PROVEAN (105), SIFT (106), or PolyPhen-2 (107), all missense variations are predicted to be damaging by at least one tool and 16 are considered damaging by all tools (Table 5). A similar result is obtained for weak D types associated with single missense variations (the 23 DEL alleles were compared with the same number of weak D alleles, which covers the range weak D type 1 to type 27, because type 4, type 11 and type 15 are not included in the ISBT weak D list and type 14 is not due to a single SNV): 2 alleles carry missense variations likely to affect splicing; all but two alleles carry variations considered deleterious by at least one tool and 15 are considered damaging by all tools. If different variations in the same codon cause either weak D or Del phenotypes, the variation causing the Del phenotype usually has a more detrimental PROVEAN score than those causing weak D or borderline weak D/Del phenotypes (e.g., RHD*01EL.10: −10.563 vs. RHD*01W.22: −9.747). However, the variation of scores observed for different codons is so large that current prediction tools must be considered insufficient to discriminate missense variations causing Del from those causing weak D phenotypes.
Table 5
| Phenotype | Allele | Protein variations | Provean (105) | SIFT (106) | PolyPhen-2 (107) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Score | Prediction | Score | Prediction | Probability | Prediction | |||||
| Del | RHD*01EL.03 | p.Leu18Pro | −5.565 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | ||
| RHD*01EL.06 | p.Leu84Pro | −4.587 | Deleterious | 0.15 | Tolerated | 1 | Probably damaging | |||
| RHD*01EL.07 | p.Ala137Glu | −2.031 | Neutral | 0.01 | Deleterious | 0.326 | Benign | |||
| RHD*01EL.10 | p.Trp408Arg | −10.563 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | |||
| RHD*01EL.12 | p.Leu153Pro | −2.036 | Neutral | 0.04 | Deleterious | 0.4 | Benign | |||
| RHD*01EL.16 | p.Gly212Arg† | −7.327 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | |||
| RHD*01EL.24 | p.Ala280Thr | −3.311 | Deleterious | 0.01 | Deleterious | 0.976 | Probably damaging | |||
| RHD*01EL.29 | p.Asp404His | −5.404 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | |||
| RHD*01EL.38 | p.Leu337Arg | −4.624 | Deleterious | 0.01 | Deleterious | 1 | Probably damaging | |||
| RHD*01EL.40 | p.Leu93Arg | −5.039 | Deleterious | 0.00 | Deleterious | 0.998 | Probably damaging | |||
| RHD*01EL.41 | p.Pro291Arg | −7.991 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | |||
| RHD*01EL.42 | p.Ser112Thr† | −2.091 | Neutral | 0.01 | Deleterious | 0.993 | Possibly damaging | |||
| RHD*01EL.43 | p.Trp16Arg | −5.567 | Deleterious | 0.18 | Tolerated | 0.71 | Possibly damaging | |||
| RHD*01EL.45 | p.Thr241Pro | −5.544 | Deleterious | 0.00 | Deleterious | 0.999 | Probably damaging | |||
| RHD*01EL.46 | p.Met295Thr | −5.422 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | |||
| RHD*01EL.49 | p.Gly339Val | −7.073 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | |||
| RHD*01EL.50 | p.Thr384Arg | −4.606 | Deleterious | 0.07 | Tolerated | 1 | Probably damaging | |||
| Del-like | RHD*09. 05 | p.[Thr201R,Phe223Val,Pro291Arg] | −7.991 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | ||
| RHD*11 | p.MetM295Ile | −3.622 | Deleterious | 0.02 | Deleterious | 1 | Probably damaging | |||
| RHD*01W.58 | p.Gly336Arg | −5.245 | Deleterious | 0.00 | Deleterious | 0.985 | Probably damaging | |||
| RHD*01W.61 | p.Arg10Trp | −6.475 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | |||
| NL-4 | p.[Thr201Arg,Phe223Val,Gly307Arg] | −3.121 | Deleterious | 0.03 | Deleterious | 0.993 | Probably damaging | |||
| NL-7 | p.Phe31Leu | −4.163 | Deleterious | 0.10 | Tolerated | 0.105 | Benign | |||
| Weak D | RHD*01W.01 | p.Val270Gly | −6.300 | Deleterious | 0.00 | Deleterious | 0.248 | Benign | ||
| RHD*01W.02 | p.Gly385Ala† | −4.861 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | |||
| RHD*01W.03 | p.Ser3Cys | −3.447 | Deleterious | 0.01 | Deleterious | 1 | Probably damaging | |||
| RHD*01W.05 | p.Ala149Asp | −4.267 | Deleterious | 0.00 | Deleterious | 0.981 | Probably damaging | |||
| RHD*01W.06 | p.Arg10Gln | −3.322 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | |||
| RHD*01W.07 | p.Gly339Glu | −6.146 | Deleterious | 0.00 | Deleterious | 0.878 | Possibly damaging | |||
| RHD*01W.08 | p.Gly307Arg | −3.121 | Deleterious | 0.03 | Deleterious | 0.993 | Probably damaging | |||
| RHD*01W.09 | p.Ala294Pro | −4.053 | Deleterious | 0.01 | Deleterious | 1 | Probably damaging | |||
| RHD*01W.10 | p.Trp393Arg | −9.563 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | |||
| RHD*01W.12 | p.Gly277Glu | −7.327 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | |||
| RHD*01W.13 | p.Ala276Pro | −4.569 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | |||
| RHD*01W.16 | p.Trp220Arg | −12.962 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | |||
| RHD*01W.17 | p.Arg114Trp | −2.610 | Deleterious | 0.02 | Deleterious | 0.999 | Probably damaging | |||
| RHD*01W.18 | p.Arg7Trp | −3.236 | Deleterious | 0.00 | Deleterious | 0.034 | Benign | |||
| RHD*01W.19 | p.Ile204Thr | 1.547 | Neutral | 0.51 | Tolerated | 0.008 | Benign | |||
| RHD*01W.20 | p.Phe417Ser | −2.753 | Deleterious | 0.00 | Few data | 0.999 | Probably damaging | |||
| RHD*01W.21 | p.Pro313Leu | −3.836 | Deleterious | 0.05 | Tolerated | 0.287 | Benign | |||
| RHD*01W.22 | p.Trp408Cys | −9.747 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | |||
| RHD*01W.23 | p.Gly212Cys† | −8.344 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | |||
| RHD*01W.24 | p.Leu338Pro | −4.381 | Deleterious | 0.01 | Deleterious | 1 | Probably damaging | |||
| RHD*01W.25 | p.Arg114Gln | −0.003 | Neutral | 0.44 | Tolerated | 0.083 | Benign | |||
| RHD*01W.26 | p.Val9Asp | −4.700 | Deleterious | 0.00 | Deleterious | 0.087 | Benign | |||
| RHD*01W.27 | p.Pro221Ser | −7.406 | Deleterious | 0.00 | Deleterious | 1 | Probably damaging | |||
†Variation probably affects splice site (102).
DEL alleles with splice site variations
The variations in many Del including the Asian-type DEL RHD*01EL.01 interfere with splicing. Usually, these variations are near the exon/intron boundary, most often in the intron. In seven alleles (ISBT: 6), the variation is 3' of the exon within 6 bp from the exon/intron junction, in six alleles (ISBT: 4), it is 5' of the exon, and in four alleles (all listed by ISBT), it is within the exon. This list may be too short as variations in the exon not adjacent to the exon/intron junction (104) or deep in the intron (108) may impact splicing.
Splice site variations may diminish or completely abolish the production of normal RHD transcripts. Incorrect splicing often results in the incorporation of intron components as pseudoexons: in RhD*01EL.08, a 19 amino acid insertion after Asn162 is predicted (62); RHD*01EL.01 transcripts often contain parts of intron 7 (21). If normal RHD transcripts are retained, normal RhD is present in the membrane and anti-D immunization of the carrier of these alleles is unlikely (102). In contrast, variations causing total absence of normal transcripts like c.486+1G>A in RHD*01EL.08 are likely to allow for anti-D immunization and to express a partial Del phenotype.
The most important example of a DEL allele with disrupted splicing is the Asian type DEL allele RHD*01EL.01. In this allele, the SNV bordering the intron/exon junction leads to exclusion of exon 9. Analysis of mRNA indicated that no transcripts with exon 9 were maintained (109). Transcripts lacking RHD exon 9 encode for a 463 amino acid protein that differs from standard RhD starting at position 385. The differences of this altered protein to the standard RhD protein are almost exclusively located in the C-terminal intracellular part of the protein. Therefore, such altered RhD protein might be able to integrate into the membrane resulting in a Del phenotype despite the lack of the ankyrin binding site. This interpretation is supported by the Del or weak D phenotype of some alleles with genomic deletions of RHD exon 9 (84) or 10 (96). However, the RHD exon 9 deletion allele leads to a partial D phenotype (84) while no anti-D observed was observed in RHD*01EL.01 probands (46). Therefore, the most likely explanation is the presence of a tiny number of normal transcripts missed by mRNA analysis. This interpretation is supported by a minigene splicing assay (102) in which full-length transcripts including RHD exon 9 were maintained in RHD*01EL.01.
Generally, minigene splicing assays (101,102) have the advantage that they allow for a systematic analysis. Still, there are limitations: the results obtained may depend on the assay used (110). Some intronic variations within the splice consensus sequence like c.148+5G>C in RHD*01EL.21 reportedly cause a Del phenotype but have limited impact in splicing assays (101). In RHD*01EL.31, the presence of normally spliced transcripts was predicted (101) but the phenotype is a partial Del (73). In conclusion, while analysis of mRNA and minigene splicing assays help to understand the impact of splice-site variations, they cannot replace serology yet. It is important that serological data are solid and confirmed by independent observations, especially if they are discrepant to the phenotype expected based on molecular studies.
DEL alleles with hybrid structure
Six alleles with hybrid structure have been correlated with a Del phenotype. Only two of these alleles (RHD*01EL.23 and RHD*01EL.44) are listed as DEL in the current ISBT table.
Typically, RhCE-like segments in RhD lead to the loss of distinct RhD epitopes (41,111). Alleles with substitutions encompassing exon 3 to exon 7 have initially been described as D− (20), those with smaller substituted segments express a distinct partial D phenotype (41,111). Often, the D antigen density of hybrid alleles is considerably reduced even if using anti-D binding to epitopes retained in these hybrids. For example, the antigen density of RHD*06.01 is about 402 antigens compared to 19,000 in a DcEe control (42). There is no direct relationship between the extent of the substitution and the antigen density (42).
Based on their structure, DEL alleles with hybrid structure are expected to express a partial Del phenotype. The lack of distinct D epitopes may lead to a misclassification as D−: use of anti-D binding to D epitopes absent in the phenotype will result in a negative adsorption/elution test. Often, only a minority of anti-Ds are binding, like 4 of 16 in RHD*01EL.44 (34). It is therefore not surprising that some alleles like RHD-RHCE(4-7)-RHD (RHD*01N.07) were initially considered D− but later repeatedly reported to underlie Del samples (34,55).
The situation is further complicated by the usually incomplete characterization of hybrid alleles: Several alleles were characterized only regarding the presence and absence of RHD exons, disregarding breakpoints or exon sequences. Therefore, it is difficult to discern if different phenotypes observed for the “same” allele in different laboratories are due to differences in the methods used for serologic characterization or due to the investigation of probands carrying different alleles.
DEL alleles with frameshift variations
At first glance, the expression of antigen D by an RHD allele with a frameshift variation is surprising. These variations are expected to lead to RhD proteins in which large protein segments do not share homology with RhD. The impact of such frameshift variations and possible mechanisms for residual D expression have recently been analyzed by Flegel and Srivastava (112): expression of D antigen was observed in 8 of 51 alleles with a frameshift variation. In five of these alleles, including four Del alleles, there were convincing explanations for the residual D expression, like the possible use of alternate start codons in alleles with variations in exon 1 (RHD*01EL.04 and RHD*01EL.18), transcriptional or translational frameshifting (RHD*01EL.18) or very limited changes in the C-terminal end of the protein with extension (RHD*01EL.11 and RHD*01EL.26). For other alleles, the Del phenotype was sometimes difficult to explain. For example, in RHD*01EL.47 a duplication of G at position 510 leads to a predicted protein of only 199 amino acids (compared to 418 in RhD) that shares only the first 171 amino acids with RhD. Still, a partial Del phenotype was reported, in which 3 of 21 anti-D were positive in the adsorption/elution test (83). For some of the remaining DEL alleles with frameshift, the Del phenotypes have never been thoroughly evaluated by serology and a D− phenotype with a false-positive adsorption/elution result cannot be excluded.
DEL alleles with premature termination codons
There are several reports of a Del phenotype expressed by alleles with a termination codon causing a truncation of the carboxy terminal intracellular end of the RhD protein [codon 401 in RHD*01EL.17 (54), codon 405 in RHD*01N.60 (11)]. Possibly, loss of the carboxy terminal part of RhD does not fully abolish antigen D expression. In contrast, the expression of antigen D by RhD proteins prematurely terminated in the middle (RHD*01EL.15 with termination at codon 308) or start (RHD*01EL.39 with termination in codon 38) of the RhD protein is more puzzling. False-positive results in adsorption/elution testing are a likely explanation for these alleles.
DEL alleles with major alterations in the RH gene including deletions of whole exons or loss of start or stop codon
RhD is part of a trimeric complex with RhAG (113), and minor alterations like missense variations in RhAG and or RhD may severely interfere with antigen D expression. Therefore, it is surprising that major alterations of the RHD gene like deletions of whole exons or loss of the normal start codon may still allow for D expression resulting in a Del phenotype.
Apart from the deletion of exon 6, each deletion of an RHD exon leads to a loss of the reading frame. However, missing exons might be replaced by pseudo-exons derived from introns. For example, in RHD*01EL.30 the 80 nucleotides of exon 8 are replaced by a 170 bp segment from intron 7 (59) retaining the reading frame for exons 9 and 10. Hence the true impact on protein structure may be less than expected.
So far, whole exon deletions in DEL alleles have been restricted to exon 1, 8, 9, and 10. Possibly, the protein segments encoded by these DNA segments are not essential for the formation of the Rh complex.
In case of RHD exon 1, there are several data hinting in this direction: (I) RHD alleles with variations in the start codon may express a Del phenotype. The most well-known example is RHD*01EL.02, the second most frequent DEL allele in many Asian populations; (II) RHD alleles with deletions of exon 1 may express a Del phenotype; (III) the bacterial RH homologue AmtB lacks the first transmembrane segment yet forms a Rh complex-like trimeric structure (114).
RHD exons 8 to 10 encode protein segments that form the last transmembrane protein segment and the intracellular tail of the RhD protein. Obviously, these segments have limited importance for the Rh complex: RHD alleles with deletions of exon 8, exon 9 or exon 10 may express RhD antigen, variations in the termination codon leading to elongated proteins often also result in a Del phenotype.
DEL alleles with exon deletions usually express a partial Del phenotype. The deletions may be triggered by homologous regions surrounding the deleted sequence: In RHD*01EL.48 (DKG), there is a 25 bp sequence identical in IVS 8 and IVS 9 (84).
Another major alteration of the RH genes is observed in NL-3: In this allele, RHCE exons 1 to 8 seem to be linked to RHD exon 9 and 10 (76), but RHD exons 1 to 7 are present yet seemingly non-expressed. The origin of the D antigen in this haplotype is unknown: RhD(1–7) seems to be unexpressed. RhCE-D(9 to 10) differs from RhCE by a single amino acid located in the C-terminal intracellular protein segment and it is difficult to imagine how such change could lead to antigen D expression.
DEL alleles with “normal” coding sequence
Repeatedly, Del phenotypes were observed in samples for which no alteration in the RHD allele could be demonstrated (26,54,66,115). In addition, three DEL alleles (RHD*01EL.32, RHD*01EL.35, and RHD*01EL.37) have initially been characterized by the presence of alterations in an intron (c.149-29G>C, c.802-41_802-38del, and c.1154-31C>T) that later turned out to represent frequent intron polymorphisms (74,77) also present in RHD alleles with normal RhD expression. Both observations suggest that the current list of possible causes of a Del phenotype is incomplete.
Worldwide distribution of Del
The distribution of DEL alleles is considerably different between East Asian and European populations (Table 6) (3,4,26,27,29-31,62,63,66,72,76,79,93,94,99,116,119,121,129,133,137,139) implicating a different importance of the Del phenotype for transfusion strategies:
Table 6
| Population | Positions screened† | Donors investigated |
D− with C or E |
D− donors screened | DEL detected | Frequency of DEL among D− |
Causative allele (individuals§) | Number of alleles‡/comments | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Argentina | 5'UTR, IVS 4, 3'UTR | 1,314 | 6 | 1:219 | RHD*01EL.43 (n=5); RHD*11 (n=1) | (80) | |||
| Argentina | 5'UTR, IVS 4, 3'UTR [no explicit description but reference to (81)] | 526 | 17 | 1:31 | RHD*01EL.43 (n=14); RHD*01EL.08 (n=1); RHD*01EL.44 (n=1); RHD*01EL.26 (n=1) | (72) | |||
| Australia | 4, 5, 10 | 2,027 | 37 (+8 DEL alleles among D−) | 1:55 | RHD*01EL.01 (n=16 incl. 2 D−); RHD*11 (n=9 incl. 1 D−); RHD*01EL.08 (n=6 incl. 1 D−); RHD*01EL.18 (n=4); RHD*01EL.09 (n=4 incl. 3 D−); RHD*01EL.05 (n=2); “RHD(ex8:del/CE)” (n=2); RHD*01EL.21 (n=1); RHD*01EL.22 (n=1); RHD*01EL.48 (n=1); RHD*01W.10 (n=1) | The molecular basis in the “RHD(ex8:del/CE)” probands was not fully resolved | (30) | ||
| Austria | 5'UTR, 3, 10 | 738 | 7 | 1:105 | RHD*11 (n=5); RHD*01EL.01 (n=1); RHD*01EL.25 (n=1) | (63) | |||
| Austria (Upper Austria ) | 4, 7, 10 | 2,427 | 3 | 1:809 | RHD*01EL.08 (n=2); RHD*09. 05 (n=1) | (88) | |||
| Austria (Upper Austria) | 4, 7, 10 | 23,330 | 66 | 1:353 | RHD*09. 05 (n=31); RHD*01EL.08 (n=24); NL-3 (n=8); RHD*01EL.04 (n=2); RHD*01EL.01 (n=1); RHD*01W.32 (n=1+1 weak D) | (94) | |||
| Bosnia-Herzegowina | 3, 5, 10 | 92 | 4 (+5 weak D) | 23 | RHD*11 (n=4);RHD*01W.01 (n=2); RHD*01W.03 (n=1); Unresolved (n=2) | (116) | |||
| Brazil | 3 | RHD*01El.01 (n=2); RHD*01EL.35 (n=1) | The algorithm used to identify the samples is not described | (117) | |||||
| Brazil | IVS 4, 10 | 239 | 0 | <1:80 | (118) | ||||
| Brazil | IVS 4, 7 | 2,450 | 10 | 1:245 | RHD*01EL.01 (n=5); RHD*09. 05 (n=5) | RHD*01EL.01 (n=5); RHD*09. 05 (n=5) | (119) | ||
| Brazil | IVS 4, 10 | 520 | 4 (+14 weak D) | RHD*01EL.01 (n=2); RHD*11 (n=1); RHD*01 (n=1) | Antigen density RHD*01EL.01: 22; RHD*11: 38; RHD: 35 | (115) | |||
| Brazil | IVS 4, 7 | 405 | 6 | 1:68 | RHD*01EL.01 (n=3); RHD*01EL.37 (n=2); RHD*01EL.32 (n=1) | RHD*01EL.01 (n=4); RHD*01EL.37 (n=2); RHD*01EL.32 (n=1) | (120) | ||
| Brazil | IVS 4, 7 | 517 | 3 | 1:172 | RHD*01EL.32 + x (n=1) RHD*01EL.01; RHD*01EL.50 | (structure of RHD*01EL.32—like sample not finally resolved); RHD*01EL.50 was not defined as Del in this study | (86) | ||
| Brazil | IVS 4, 7 | 1,403 | 1 | 1:1,403 | NL-9 (n=1) | (86) | |||
| Canada | 3,980 | RHD*01EL.18 (n=7) | Allele frequency mentioned in the introduction | (121) | |||||
| China | 1, 4, 5, 6, 7 | 102 | 26 | RHD*01EL.01 (n=25); RHD*01EL.36 (n=1) | RHD*01EL.01 (n=34); RHD*01EL.36 (n=1) | (15) | |||
| China | 3, 4, 5, 6, 7, 9 | 74 | 22 | 1:3.4 | RHD*01EL.01 (n=22) | RHD*01EL.01 (n=22) | (122) | ||
| China | Adsorption/elution; 3, 4, 5, 6, 7, 9 | 15,643 (Han: 12,546; Uigur: 1,814) | 150 (Han: 50; Uigur: 86) | 14 (Han: 11; Uigur: 2) | 1:11 (Han: 1:4.5; Uigur: 1:43) | RHD*01EL.01 (n=14) | (123) | ||
| China | Adsorption/elution | 400,253 | 1,585 | 279 | 1:5.7 | RHD*01EL.01 (n=268); RHD*01EL.02 (n=4); RHD*01EL.44 (n=1); RHD*01EL.03 (n=1); RHD*01EL.06 (n=1); RHD*01EL.07 (n=1); RHD*01W.61 (n=1); NL-6 (n=1); NL-8 (n=1) | (3) | ||
| China (Han) Hefei | 5'UTR, IVS 4, 7, 10 | 152 | 31 | 1:4.9 | RHD*01EL.01 (n=31) | (124) | |||
| China | Testing for RHD*01EL.01; RHD all exons | 2,493 | 516 | 1:4.8 | RHD*01EL.01 (n=516) | RHD*01EL.01 (n=565) | (24); overlap with (125) | ||
| China | Testing for RHD*01EL.01 | 2,385 | 516 | 1:4.6 | RHD*01EL.01 (n=516) | RHD*01EL.01 (n=565) | (125); overlap with (24) | ||
| China | Serologic testing by adsorption/elution | 165 | 41 | 1:4.0 | RHD*01EL.01 (n=37); RHD*01EL.24 (n=1); RHD*01N.07 (n=1); NL-7 (n=1); NL-8 (n=1) | RHD*01EL.01 (n=39); RHD*01EL.24 (n=1); RHD*01N.07 (n=1); NL-7 (n=2); NL-8 (n=1) | (55) | ||
| China | Testing for RHD*01EL.01; serologic testing by adsorption/elution | 643 | 155 | 1:4.1 | RHD*01EL.01 (n=151); other (n=4) | (33) | |||
| China | Serologic testing by adsorption/elution | 808 | 178 | 1:4.5 | RHD*01EL.01 (n=158); RHD*01EL.02 (n=8); RHD*01EL.44 (n=1); NL-8 (n=1) | (35) | |||
| China | 804; subset: 515 pregnant women |
221; subset: 142 |
Molecular analysis only for subset: RHD*01EL.01 (n=130); RHD*01EL.44 (n=7); NL-8 (n=3); RHD*01N.07 (n=1) | (34) | |||||
| China (Taiwan) | RT-PCR and RFLP? | 204 | 41 | NL-1 or RHD*01EL.01 (n=41) | Allele reported as NL-1 but data compatible with RHD*01EL.01 | (126) | |||
| China (Taiwan) | serologic testing by adsorption/elution; 2, 3, 5, 7, 9 | 156 | 34 | 1:4.6 | exon 2,3,5,7,9 detected (n=27: 26 C+E-, 1 C-E-); exon 2,3,5,7 detected (n=7; all C+E-) | structure of alleles not resolved beyond PCR pattern; PCR patterns observed in Del and D− difficult to reconcile with other studies | (127) | ||
| China (Taiwan) | Serologic testing by adsorption/elution; 4, 5, 7, 9, 10; Testing for RHD*01EL.01 | 294 | 94 | RHD*01EL.01 (n=94) | RHD*01EL.01 (108) | (16) | |||
| China (Taiwan) | Testing for RHD*01EL.01; Adsorption/elution | 395 | 130 | 1:3.0 | RHD*01EL.01 (n=126 + 1 D−); other (4) | One RHD*01EL.01 sample was D− | (25) | ||
| China (Taiwan) | Serologic testing by adsorption/elution; Testing for RHD*01EL.01 | 118 | 38 | RHD*01EL.01 (n=38); + 1 RHD*01EL.01-like among D− | (18) | ||||
| Croatia | Serologic testing with IAT | 1,630 | 6 | 1:272 | RHD*11 (n=6) | Adsorption/elution only done in samples faintly positive in IAT; antigen density 28 to 44 (median 35) | (128) | ||
| Croatia | 7, 10 | 6,523 | 12 | 1:544 | RHD*11 (n=4); RHD*01 (n=4); RHD*01W.02 (n=2); RHD*01W.28 (n=1); NL-10 (n=1) | RHD*01W.02 and RHD*01W.28 samples had a “C” in trans. The molecular cause of Del phenotype in RHD*01 remained unresolved despite sequencing all exons | (99) | ||
| Denmark | 10 | 233 | 3 | 1:78 | RHD*01EL.08 (n=2); RHD*01EL.01 (n=1); RHD*01EL.02 (n=1) | (62) | |||
| Denmark | 5, 7, 10 | 5,058 (4,932 results available) | 2 | 1:2,029 | RHD*01EL.33 (n=2); RHD*01EL.08 (n=1 D−) | The RHD*01EL.08 sample was considered D− | (53) | ||
| Finland | 5, 7 | 16,253 | 5; +5 possible | 1:3,250 (to 1:1,625) | RHD*01EL.01 (n=2); RHD*01EL.04 (n=1); RHD*01EL.08 (n=1); RHD*11 (n=1); possible DEL: RHD*c.829G>A (n=4); RHD*c.[1154G>C;1163T>G] (n=1) | “Possible Del”: negative in routine serology but molecular variation atypical for D− | (129) | ||
| Germany (South-West) | 5'UTR, IVS 4, 7, 10 | 754 | 15 (+4 weak D/partial D) |
1:50 | RHD*11 (n=7); RHD*01EL.01 (n=5); RHD*01EL.08 (n=3) | (20) | |||
| Germany (South-West) | 5'UTR, IVS 4, 10 | 7,688 | 0 | <1:2,500 | (20) | ||||
| Germany (North) | 5'UTR, 3, 10 | 454 | 2 | 1:227 | RHD*11 (n=2) | (63) | |||
| Germany (South-West) | IVS 4 | 46,133 | 47 | 1:982 | RHD*01EL.08 (n=16); RHD*11 (n=14); RHD*01EL.01 (n=4); RHD*01EL.04 (n=4); RHD*01EL.18 (n=2); RHD*01EL.25 (n=2); RHD*01EL.12 (n=1); RHD*01EL.16 (n=1); RHD*01EL.17 (n=1); RHD*01EL.23 (n=1); RHD*01 (n=1) | (54) | |||
| India | Serologic testing by adsorption/elution | 200 | 3 | 1:67 | 2 Del C+, 1 Del E+ | (130) | |||
| India | Serologic testing by adsorption/elution; molecular testing for RHD*01EL.01 and RHD*11 | 900 | 0 | <1:243$ | (131) | ||||
| India | Serologic testing by adsorption/elution | 1,003 | 2 | 1:502 | Both Del C+ | (132) | |||
| Italy | Commercial PCR systems, pools of 5 | 235 | 1 | 1:235 | RHD*11 (n=1) | (133) | |||
| Japan | 1, 2, 3, 4–6, 7, 8, 9, 10 | 3,526 | 324 | 1:11 | RHD*01EL.01 (n=318); RHD*01 (n=3); RHD*01EL.25 (n=2); RHD*01EL.08 (n=1) | RHD*01EL.01 (n=329); RHD*01 (n=3); RHD*01EL.25 (n=2); RHD*01EL.08 (n=1) | (26) | ||
| Japan | 1 to 10 | 2,754 | 240 | RHD*01EL.01 (n=232); RHD*01EL.25 (n=2); RHD*01EL.10 (n=1); RHD*01EL.45 (n=1); RHD*01N.60 (n=1); RHD*01N.70 (n=1); Unresolved (n=2) | (11) | ||||
| Korea | IVS 4, 7 | 126 | 16 | 1:7.9 | RHD*01EL.01 (n=16) | RHD*01EL.01 (n=16) | (17) | ||
| Korea | 3, IVS 4, 5,7,10 | 264 | 43 | 1:6.1 | RHD*01EL.01 (n=42); RHD*01EL.10 (n=1) | RHD*01EL.01 (n=42); RHD*01EL.10 (n=1) | (27) | ||
| Korea | Promoter, IVS 4, 7, 10 | 110 (“D-negative club” members) | 16 | 1:6.9 | RHD*01EL.01 (n=14); RHD*01EL.10 (n=2) | (134) | |||
| Korea | 95 | 17 | 1:5.6 | RHD*01EL.01 (n=17) | (135) | ||||
| Korea | Promoter, IVS 4, 7, 10 | 50 | 27 | 1:1.9 | RHD*01EL.01 (n=26); RHD*01EL.10 (n=1) | (136) | |||
| Morocco | Serologic testing by adsorption/elution | 425 | 4 | (137) | |||||
| Myanmar | Serologic testing by adsorption/elution | 222 | 35 | 1:6.3 | No molecular characterization | 9 weak D found | (138) | ||
| Netherlands | 5, 7 | 37,782 | 34 | 1:1,111 | RHD*01EL.01 (n=9); RHD*01EL.45 (n=7); RHD*01EL.18 (n=6); NL-4 (n=3); RHD*(1-9) (n=3); RHD*01 (n=3); RHD*01EL.11 (n=1); RHD*01EL.17 (n=1); RHD*01EL.46 (n=1) | RHD*01EL.01 (n=9); RHD*01EL.45 (n=7); RHD*01EL.18 (n=6); NL-4 (n=4); RHD*(1-9) (n=3) RHD*01 (n=3); RHD*01EL.11 (n=1); RHD*01EL.17 (n=1); RHD*01EL.46 (n=1) | (66) | ||
| Poland | IVS 4, 7, 10 | 31,200 | 10 | 1:3,120 | RHD*11 (n=5); NL-6 (n=3); RHD*01EL.08 (n=1); RHD*01EL.32 (n=1) | (76) | |||
| Russia | 5'UTR, 3, 10 | 71 | 0 | <1:19$ | (63) | ||||
| Serbia | 3, 5, 10 | 61 | 5 | 1:12 | RHD*11 (n=2); RHD*15 (n=2); RHD*01W.02 (n=1) | (116) | |||
| Slowenia | 5'UTR, 3, 10 | 333 | 4 | 1:83 | RHD*01EL.01 (n=3); RHD*11 (n=1) | (63) | |||
| Switzerland | 5'UTR, 3, 10 | 104 | 2 | 1:52 | RHD*01EL.01 (n=2) | (63) | |||
| Switzerland (BTS Berne) | 3, 5, 10 | 20,015 | 27 | 1:741 | RHD*11 (n=10); RHD*01EL.08 (n=6); RHD*01EL.46 (n=3); RHD*01EL.01 (n=2); RHD*01EL.18 (n=1); RHD*01EL.41 (n=1); RHD*01EL.42 (n=1); RHD*01N.48 (n=1); NL-2 (n=1); RHD*01EL.43 (n=1) | Donors tested 2012; RHD*11 and RHD*01EL.43 weakly positive in IAT but considered DEL | (79); dataset possibly overlapping with (93) | ||
| Switzerland (BTS Zurich) | 5,7,10 | 5,355 | 5 | 1:1,071 | RHD*11 (n=2) RHD*01EL.08 (n=1); NL-5 (n=2) | Donors tested 2012; RHD*11 weakly positive in IAT but considered DEL | (79); dataset possibly overlapping with (93) | ||
| Switzerland (BTS Berne) | 3, 5, 10 | 652 | 9 | 1:72 | RHD*11 (n=4); RHD*01EL.08 (n=3); NL-5 (n=2) | (93); dataset possibly overlapping with (79) | |||
| Switzerland (BTS Berne) | 3, 5, 10 | 17,391 | 1 | 1:17,391 | RHD*01EL.08 (n=1) | (93); dataset possibly overlapping with (79) | |||
| Switzerland (BTS Zurich) | IVS 4, 7 | 8,200 | 10 | 1:820 | RHD*11 (n=5); RHD*01EL.08 (n=4); RHD*01EL.01 (n=1) | (93); dataset possibly overlapping with (79) | |||
| Thailand | (Serologic screening) | 254 | 50 | 1:5.1 | RHD*01EL.01 (n=48); other (n=2) | (28) | |||
| Thailand | 1 to 10 | 321 | 121 | 1:2.6 | RHD*01EL.01 (n=108+21*); RHD*01N.01 (n=11); RHD*01N.03 (n=1); RHD*15 (n=1) | *21 RHD*01EL.01 alleles were found in “D-negative” samples, vice versa 11 “Del” samples were homozygous for the RHD deletion | (32) | ||
| Thailand | 1 to 10 | 1,125 | 180 | 1:6.3 | RHD*01EL.01 (n=175); RHD* 01EL.44 (n=4); RHD*01EL.08 (n=1). Three additional alleles were detected in a single sample and reported as D− without testing by adsorption/elution RHD*c.325A>C; RHD*c.604G>A; RHD*c.[1136C>T;1223G>A] | RHD*01EL.01 (n=186); RHD* 01EL.44 (n=5); RHD*01EL.08 (n=1) | (29) | ||
| Tunisia | 10 | 488 | 4 (incl. weak D/partial D) | 1:122 | RHD*11 (n=2); RHD*09.03.01 (n=1); RHD*01W.29 (n=1) | (139) | |||
| USA | Variant testing using beadchip assay | 1,174 | 6 | 1:196 | RHD*01EL.01 (n=4); RHD*01EL.08 (n=1); RHD*01EL.09 (n=1) | (31) |
†Numbers indicate exons tested by PCR. §Number of individuals with this allele (i.e., homozygous occurrence counted as 1). ‡Number of alleles (i.e., homozygous occurrence counted as 2). Numbers only indicated if different to number of individuals. *In addition, an RHD*01EL.08 sample was detected but characterized as D-negative. $Upper limit of 95% confidence interval (binomial distribution), estimated frequency 0. IVS, intervening sequence; UTR, untranslated region; incl., including; IAT, indirect antiglobulin test.
In typical East Asian populations, donors D− by routine serology are rare and usually comprise less than 1% of all donors [e.g., China: 0.4% (3,123) to 1% (140), Japan 0.5% (141)]. In China, about 24% of these donors have a Del phenotype (Japan, 9%; Korea, 15%; Thailand, 20%; percentages are median results of the surveys in Table 6). In 97% (China, Japan, Thailand) to 99% (Korea) of Del phenotypes the underlying allele is the Asian-type RHD*01EL.01. Hence, definition of the transfusion strategy for one single DEL allele, RHD*01EL.01, has major impact for transfusion support of patients typed D− by routine serology.
In contrast, in European populations, only 0.03% (Poland) to 0.28% (Croatia) of probands who are D− by routine serology possess a Del phenotype. Furthermore, the underlying alleles are heterogeneous and often not suitable for D+ transfusion. As a result, there is no advantage of identifying Del in patients. A possible exception may be patients with c or e negative phenotypes for whom Rh phenotype compatible D− blood is difficult to provide even in Europe. Still, the major Del issue in these populations is a possible anti-D immunization risk incurred by Del donors mistyped as D−.
As a corollary, it should be noted that the “European-derived” situation is not as uniform as it might seem: On closer inspection, relevant differences of the frequency of the “rare” DEL alleles can be detected. RHD*01EL.08 is frequent in Central Europe [Austria (94), Denmark (62), Germany (20), Switzerland (79,93)]. RHD*01EL.18 is found in francophone Canadians (121). RHD*01EL.43 is dominant in Argentina (72) but otherwise rare. RHD*11 is frequent in Central (20,54,63,76,79,93) and South-Eastern (116,128) Europe. RHD*09.05 is frequent in parts of Austria (94) and in Brazil (119) in donors of African descent but seems to be rare elsewhere. In Austria, there is an obvious founder effect: Most carriers of RHD*09.05 live near the river Traun (142) and possess a RHCE*ce.20.13 allele. As RHD*09.05 is one of the few Del types that can be found among seemingly D− C-E- samples, such local variation may be of importance for typing strategies.
Anti-D immunization
Anti-D immunization in Del patients
Anti-D immunization is unlikely in patients with the RHD*01EL.01 allele (see paragraph “Asian-type DEL”). However, anti-D antibodies have been observed in patients with other DEL alleles (Table 7) (34,35,43,59,60,73,143).
Table 7
| Allele | Observation | Reference |
|---|---|---|
| RHD*01EL.04 | Anti-D observed in pregnancy | (60) |
| RHD*01EL.08 | 2 females with anti-D | (43) |
| RHD*01EL.08 | 2 mothers with anti-D; mild HDN in one child | (143) |
| RHD*01EL.30 | Woman (2 stillbirth) with anti-D | (59) |
| RHD*01EL.31 | Anti-D causing severe hemolytic disease in 2nd child | (73) |
| RHD*01EL.44 | Anti-D observed in 3 of 7 individuals in survey | (34) |
| RHD*01EL.44 | Anti-D observed in the single individual in survey | (35) |
| RHD*01N.07 | Anti-D observed in the single individual in survey | (34) |
| NL-8 | Anti-D observed in 2 of 4 individuals in survey | (34) |
| NL-8 | Anti-D observed in the single individual in survey | (35) |
Most of these anti-D were detected in pregnant females with hemolytic disease of the fetus or newborn, which can be severe (73). Anti-D immunizations have been reported for RHD*01EL.04, RHD*01EL.08, RHD*01EL.30, RHD*01EL.31, RHD*01EL.44, RHD*01N.07, RHD*11 and NL-8. These alleles must therefore be considered partial Del. Three of them (RHD*01EL.44, RHD*01N.07, and NL-8) are RHD/RHCE hybrid alleles, two (RHD*01EL.08 and RHD*01EL.31) carry splice site variations at position +1, the two remaining alleles are due to a frameshift in exon 1 (RHD*01EL.04) and the deletion of RHD exon 8 (RHD*01EL.30), respectively. Additional Del for which a partial Del phenotype has been predicted are RHD*01EL.09, RHD*01EL.19, RHD*01EL.22, RHD*01EL.33, RHD*01EL.47, and anti-D immunization is likely possible in carriers of these alleles. The same is true for carriers of alleles that are likely D− rather than Del.
The prediction of a partial Del phenotype based on its structure is sometimes difficult. Even the observation of an anti-D may be misleading: Even more than in weak D, the discrimination of an allo-anti-D from an auto-anti-D is difficult in Del: due to the low antigen density, a high titer auto-anti-D may be associated with a negative direct antiglobulin test. An example of auto-anti-D has been reported for RHD*11 (81). Hence, the absence of an anti-D immunization risk in carriers of a specific DEL allele must be based on the observation of a large number of Del individuals. Despite several surveys (Table 2), convincing data are available only for RHD*01EL.01 although the database is expected to vastly improve over time for the Asian-type and many other DEL alleles.
Anti-D immunization of D− patients by RBC units from Del donors
When the molecular investigation of seemingly D− blood donors revealed the presence of donors expressing D in the D− donor pool (20), the issue of possible anti-D immunization of D− patients by Del blood units was raised immediately and a look back study was included in this publication. The first observation of an anti-D immunization caused by Del units was a female Austrian patient who was shown to have received an RHD*01EL.35 unit (4). Shortly afterwards a considerable increase of the anti-D titer in a pre-immunized 67 years old Japanese woman receiving two RHD*01EL.01 units was reported (144). The latter observation was important because RHD*01EL.01 is the by far most frequent DEL allele in Asians (15-18).
Examples of possible anti-D immunization events caused by blood donations from Del blood donors are summarized in Table 8 (4,11,97,121,144-150). Most reports concern RHD*01EL.01, which is not surprising since it is by far most frequent DEL allele.
Table 8
| Allele | Type | Sex | Age (years) | Observation | Reference | |
|---|---|---|---|---|---|---|
| RHD*01EL.01 | S | F | 67 | Two RHD*01EL.01 units among 59 “D neg” units. Titer increase <1 to 8 after 1st unit; 8 to 128 after 2nd Del unit | (144) | |
| RHD*01EL.01 | P | M | 68 | 1 RHD*01EL.01 unit among 4 units transfused; anti-D 9 days after transfusion | (145) | |
| RHD*01EL.01 | P | M | 68 | 2 DEL units -> new anti-D titer 2 after 22 days | (146) | |
| S | F | 33 | 1 DEL unit -> titer increase 8 to 64 | |||
| S | M | 45 | 2 DEL units -> titer increase 8 to 64 | |||
| RHD*01EL.01 | P | M | 64 | New anti-D 5 to 7 d after transfusion of 2 Del unit from 2 donors | (147) | |
| RHD*01EL.01 | P | F | 54 | new anti-D 2 month after transfusion of 1 RHD*01EL.01 unit | (148) | |
| RHD*01EL.01 (probable) | P | M | 64 | New anti-D 1 month after transfusion of 2 Del units | (149) | |
| S | M | 73 | New anti-D 6 days after transfusion of 4 Del units | |||
| RHD*01EL.01 (probable) | S | F | 70ies | “new” anti-D 34 days after 1 unit RHD*01EL.01 | Compiled in (11) | |
| S | F | 57 | Titer <1 → 4096; first detected 2 years after 2 unit RHD*01EL.01 | |||
| S | F | 70ies | “new” anti-D + anti-C 28 days after 1 unit C+ | |||
| S | F | 85 | Titer <1 → 8; first detected day 4 after 1 unit C+ | |||
| P | M | 35 | New anti-D + Anti-C 5 months after 1 unit DEL | |||
| S | M | 79 | Titer <1 → 8, first detect 4 days after 1 unit RHD*01EL.01 | |||
| S | F | 86 | New anti-D 9 days after 1 unit C+ | |||
| S | F | 80ies | Titer <1 → 2 2 weeks after 1 unit RHD*01EL.01 | |||
| RHD*01EL.18 | F | 87 | Anti-D detected (see also Table 9) | (121) | ||
| F | 88 | Anti-D detected (see also Table 9) | ||||
| F | 44 | Anti-D detected (see also Table 9) | ||||
| F | 88 | Anti-D detected (see also Table 9) | ||||
| RHD*01EL.35 | P | F | 58 | New anti-D 8 days after transfusion of 1 RHD*01EL.35 unit | (4) | |
| NL-5 | S | F | 76 | New anti-D 10 days after transfusion of 1 NL-5 unit | (97) | |
| RHD*01 | P? | M? | ? | Anti-D 45 days after transfusion of 1 DEL unit; molecular analysis of the DEL detected no difference from RHD*01 | (150) | |
P, primary immunization; S, secondary immunization; F, female; M, male; 70ies, age 70 to 79; 80ies, age 80 to 89; ?, not detailed or information not clearly described.
Considering the many reports accrued, there is no doubt that Del RBC units may cause anti-D immunization. The evidence for primary anti-D immunization by Del is less strong: There are 8 reports suggesting a primary anti-D formation after transfusion of Del units, but in five of these, the anti-D occurred within one month. Although such rapid primary anti-D immunization is possible, it is suspicious: in most studies on D+ transfusion of D− probands, the anti-D occurred after 2 to 9 months with a mean of 17 weeks in one study (151,152). This finding was replicated in D− trauma patients after D+ transfusion in whom no anti-D formation was detected before 3 months and 8 of 9 anti-D were detected after more than 6 months (153). The “rapid” immunization by Del RBC units might be an observation bias (anti-D shortly after a D− transfusion is likely to trigger further investigations) but could also indicate that the assumed “primary” immunization represents a secondary immune response. In addition, in several reports, transfusion of D+ platelet units represents a possible alternative cause of anti-D immunization.
The likelihood of anti-D production has been targeted in several studies investigating the fate of patients transfused with Del RBC units (Table 9) (31,33,53,63,121,146,150). Usually only a few transfusions were informative: Many patients receiving Del RBC units were D+, had preexisting anti-D or were lost to follow-up. If only D− patients with follow up are considered, there were 103 informative transfusions among which 6 anti-D immunizations occurred. Four of these immunization events could alternatively have been caused by D+ platelet units. The risk of anti-D immunization after transfusion with “standard” D+ units is 20% to 50% depending on the patient population (151,152,154). An immunization risk of 4% to 7% would thus be about one tenth of the risk incurred by a normal D+ unit.
Table 9
| Survey | Country | Allleles | Anti-D* | Follow-up | Comment |
|---|---|---|---|---|---|
| Gassner et al. 2005 (63) | Austria | RHD*01EL.11 | 0/1 | Not reported | |
| RHD*11 | 0/7 | 17 to 145 days | |||
| Krog et al. 2011 (53) | Denmark | RHD*01EL.08 | 0/11 | <3 months (5) to >12 months (3) | D pos platelet units in 1 of 2 immunized patients |
| RHD*01EL.33 | 2/29 | <3 months (14) to >12 months (8) | |||
| Shao et al. 2012 (146) | China | RHD*01EL.01 | 1/6 | Not reported | 3 additional patients with pre-existing anti-D: anti-D boostered in 2 of 3; insufficient Hb increase |
| St-Louis et al. 2013 (121) | Canada | RHD*01EL.18 | 3/31 | D pos platelet units | |
| RHD*11 | 0/1 | ||||
| Wang et al. 2014 (33) | China | RHD*01EL.01 | 0/14 | Not reported | |
| Perez-Alvarez et al. 2019 (31) | USA | RHD*01EL.01 | 0/1 | 37 days | |
| RHD*01EL.08 | 0/2 | 711 to 729 days | |||
| Safic Stanic et al. 2022 (150) | Croatia | RHD*01 | 1/? | Not reported | The number of informative transfusions is not reported per allele. In total, there were 40 informative D− recipients |
| RHD*11 | 0/? | ||||
| RHD*01W.2 | 0/? | ||||
| RHD*01W.28 | 0/? | ||||
| NL-10 | 0/? |
*Number of observed anti-D immunization and number of D− patients without anti-D prior to transfusion transfused and not lost to follow up. ?, data not disclosed in the reference.
Some authors (53) suggest that such low immunization rate does not support general testing of blood donors for Del, especially in a population where avoiding C and E positive units is an alternative strategy (155). The situation is similar to weak D where cases of anti-D immunization by weak D units are well documented but this event is rare in prospective observations (156). Sometimes, the low risk of anti-D immunization is reinforced by comparing the immunization rates to those caused by other blood group antigens. Such comparisons miss an important point: despite the low immunogenicity of Del units, anti-D remains the leading cause of severe hemolytic disease of the fetus in many populations (157,158) and preexisting anti-D-immunization, even if caused by a Del unit, makes anti-D prophylaxis futile. Thus, in many regions the harm caused by an anti-D immunization is more than that caused by an anti-E immunization.
An additional argument against the use of Del units for D− patients is a reduced hemoglobin increase after transfusion of Del units to patients with high titer anti-D observed by Shao et al. (146).
Some blood services started to identify Del donors by RHD PCR of blood donors [serologic testing is less suitable (58)]. Three approaches have been considered [also reviewed in (5)]:
(I) The most straightforward approach consists of testing all D− donors for Del. This algorithm is the “safest” one, as it does not depend on any assumptions regarding haplotype association. On the downside, it necessitates more testing than the other algorithms. This algorithm is obligatory in Switzerland (79), listed as possible approach in the German transfusion guidelines (159) and has been implemented by several blood services in Austria (94), Germany (54) and the United States (31). As prices for molecular techniques are generally decreasing while those for serologic testing tend to increase, it might even become cost efficient if it replaces sensitive serologic testing for D in blood donors (5). Still, the costs reported for 2012 in Switzerland were between 7 and 10 € per sample investigated (79) which is considerably higher than serologic testing by IAT.
(II) Since the vast majority of DEL alleles occur in a DCe or DcE haplotype, algorithms have been proposed that limit testing for Del to D−, C+ probands (25,28,58,134) or to D− probands whose RBC express C or E. In many countries, determination of the full Rh phenotype is routine and such algorithm may reduce testing considerably. This approach has been advocated in China (including Taiwan) (25), Korea (134), and Thailand (28). It should be noted, however, that the algorithm will fail in populations in which RHD*09.05 or other ce-associated DEL alleles are frequent, like Austria and Argentina. Furthermore, there are rare observations of RHD*01EL.01 in C negative probands (15).
(III) Finally, it has been discussed that transfusion patients at high risk of anti-D immunization exclusively with D− RBC units lacking antigen C (11) or lacking antigens C and E (155) might be an alternative to testing for Del. Obviously, such strategy is easier in countries with plenty of D− units lacking C and E than in East Asian populations where D− units are scarce and often carry antigen C.
Conclusions
Much progress has been made in defining the molecular basis of Del and the serological and clinical implications of this phenotype. More than 45 distinct DEL alleles are recognized by the ISBT, ten additional alleles might also qualify as DEL. The most frequent mechanisms leading to DEL alleles are missense variations and variations interfering with splicing. Less frequently, hybrid alleles, frameshift variations, premature termination codons and large deletions encompassing whole exons are observed. The current knowledge has relevant limitations: Some variations observed in DEL alleles cannot influence the phenotype and in some Del samples no variations were detected. Technical confounders include an ill-defined boundary between weak D and Del as well as false-positive and false-negative adsorption/elution results impeding the serologic discrimination of Del from D negative. Similar to weak D alleles, distinct DEL alleles display distinct serologic profiles that differ in the RhD antigen density, the absence of D epitopes and a possible anti-D immunization risk in carriers of these DEL alleles. The prevalence of DEL alleles varies widely between populations, resulting in different implications for transfusion strategies: In East Asian populations, a relevant proportion of individuals D− in routine serological D testing possess the RHD*01EL.01 allele. Hence, in these populations, definition of the correct transfusion strategy for patients with RHD*01EL.01 has relevant impact on the D− blood supply, even more than the D+ transfusion strategy for the frequent weak D types in Europe. In contrast, in many other populations, Del is a rarity and caused by numerous different alleles. In such populations, the main focus is the prevention of anti-D immunization of D− patients by Del blood units released as D negative.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Lilian Castilho) for the series “Serology and Molecular Biology of the Rh System” published in Annals of Blood. The article has undergone external peer review.
Reporting Checklist: The author has completed the Narrative Review reporting checklist. Available at https://aob.amegroups.com/article/view/10.21037/aob-22-16/rc
Peer Review File: Available at https://aob.amegroups.com/article/view/10.21037/aob-22-16/prf
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://aob.amegroups.com/article/view/10.21037/aob-22-16/coif). The series “Serology and Molecular Biology of the Rh System” was commissioned by the editorial office without any funding or sponsorship. FFW reports royalties for patents on RHD molecular biology (patents ended in 2020). FFW is a member of the blood group database advisory board of ISBT and a member of the working party on blood group antigen terminology of ISBT. FFW administrates the Human RhesusBase. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Peyrard T, Wagner F. The Rh System. In: Cohn CS, Delaney M, Johnson ST, et al. editors. Technical Manual. 20 ed. AABB, 2020:329-54.
- Okubo Y, Yamaguchi H, Tomita T, et al. A D variant, Del? Transfusion 1984;24:542. [Crossref] [PubMed]
- Li Q, Hou L, Guo ZH, et al. Molecular basis of the RHD gene in blood donors with DEL phenotypes in Shanghai. Vox Sang 2009;97:139-46. [Crossref] [PubMed]
- Wagner T, Körmöczi GF, Buchta C, et al. Anti-D immunization by DEL red blood cells. Transfusion 2005;45:520-6. [Crossref] [PubMed]
- Wagner FF. RHD PCR of D-Negative Blood Donors. Transfus Med Hemother 2013;40:172-81. [Crossref] [PubMed]
- Nuchnoi P, Thongbus J, Srisarin A, et al. Clinical and laboratory update on the DEL variant. Lab Med 2014;45:285-90. [Crossref] [PubMed]
- Kwon DH, Sandler SG, Flegel WA. DEL phenotype. Immunohematology 2017;33:125-32. [Crossref] [PubMed]
- Sandler SG, Flegel WA. Does transfusion of Asian-type DEL red blood cells to D- recipients cause D alloimmunization? Transfusion 2019;59:2455-8. [Crossref] [PubMed]
- Flegel WA, Wagner FF. DEL. Blood Transfus 2020;18:159-62. [PubMed]
- Yin Q, Flegel WA. DEL in China: the D antigen among serologic RhD-negative individuals. J Transl Med 2021;19:439. [Crossref] [PubMed]
- Ito S, Ohto H, Ogiyama Y, et al. A practical and effective strategy in East Asia to prevent anti-D alloimmunization in patients by C/c phenotyping of serologic RhD-negative blood donors. EJHaem 2021;2:750-6. [Crossref] [PubMed]
- Wagner FF, Flegel WA. The Rhesus Site. Transfus Med Hemother 2014;41:357-63. [Crossref] [PubMed]
- Floch A, Téletchéa S, Tournamille C, et al. A Review of the Literature Organized Into a New Database: RHeference. Transfus Med Rev 2021;35:70-7. [Crossref] [PubMed]
- Mak KH, Yan KF, Cheng SS, et al. Rh phenotypes of Chinese blood donors in Hong Kong, with special reference to weak D antigens. Transfusion 1993;33:348-51. [Crossref] [PubMed]
- Shao CP, Maas JH, Su YQ, et al. Molecular background of Rh D-positive, D-negative, D(el) and weak D phenotypes in Chinese. Vox Sang 2002;83:156-61. [Crossref] [PubMed]
- Chen JC, Lin TM, Chen YL, et al. RHD 1227A is an important genetic marker for RhD(el) individuals. Am J Clin Pathol 2004;122:193-8. [Crossref] [PubMed]
- Luettringhaus TA, Cho D, Ryang DW, et al. An easy RHD genotyping strategy for D- East Asian persons applied to Korean blood donors. Transfusion 2006;46:2128-37. [Crossref] [PubMed]
- Sun CF, Liu JP, Chen DP, et al. Use of real time PCR for rapid detection of Del phenotype in Taiwan. Ann Clin Lab Sci 2008;38:258-63. [PubMed]
- Shao CP. Transfusion of RhD-positive blood in "Asia type" DEL recipients. N Engl J Med 2010;362:472-3. [Crossref] [PubMed]
- Wagner FF, Frohmajer A, Flegel WA. RHD positive haplotypes in D negative Europeans. BMC Genet 2001;2:10. [Crossref] [PubMed]
- Liu HC, Eng HL, Yang YF, et al. Aberrant RNA splicing in RHD 7-9 exons of DEL individuals in Taiwan: a mechanism study. Biochim Biophys Acta 2010;1800:565-73. [Crossref] [PubMed]
- Xiong W, Shao CP, Li XM, et al. Research of RhD protein in Rh blood group Del phenotype. Zhonghua Yi Xue Za Zhi 2006;86:128-32. [PubMed]
- Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434-43. [Crossref] [PubMed]
- Ye SH, Wu DZ, Wang MN, et al. A comprehensive investigation of RHD polymorphisms in the Chinese Han population in Xi'an. Blood Transfus 2014;12:396-404. [PubMed]
- Wang YH, Chen JC, Lin KT, et al. Detection of RhD(el) in RhD-negative persons in clinical laboratory. J Lab Clin Med 2005;146:321-5. [Crossref] [PubMed]
- Ogasawara K, Suzuki Y, Sasaki K, et al. Molecular basis for D- Japanese: identification of novel DEL and D- alleles. Vox Sang 2015;109:359-65. [Crossref] [PubMed]
- Kim JY, Kim SY, Kim CA, et al. Molecular characterization of D- Korean persons: development of a diagnostic strategy. Transfusion 2005;45:345-52. [Crossref] [PubMed]
- Srijinda S, Suwanasophon C, Visawapoka U, et al. RhC Phenotyping, Adsorption/Elution Test, and SSP-PCR: The Combined Test for D-Elute Phenotype Screening in Thai RhD-Negative Blood Donors. ISRN Hematol 2012;2012:358316. [Crossref] [PubMed]
- Thongbut J, Laengsri V, Raud L, et al. Nation-wide investigation of RHD variants in Thai blood donors: Impact for molecular diagnostics. Transfusion 2021;61:931-8. [Crossref] [PubMed]
- Scott SA, Nagl L, Tilley L, et al. The RHD(1227G>A) DEL-associated allele is the most prevalent DEL allele in Australian D- blood donors with C+ and/or E+ phenotypes. Transfusion 2014;54:2931-40. [Crossref] [PubMed]
- Perez-Alvarez I, Hayes C, Hailemariam T, et al. RHD genotyping of serologic RhD-negative blood donors in a hospital-based blood donor center. Transfusion 2019;59:2422-8. [Crossref] [PubMed]
- Thongbut J, Raud L, Férec C, et al. Comprehensive Molecular Analysis of Serologically D-Negative and Weak/Partial D Phenotype in Thai Blood Donors. Transfus Med Hemother 2020;47:54-60. [Crossref] [PubMed]
- Wang QP, Dong GT, Wang XD, et al. An investigation of secondary anti-D immunisation among phenotypically RhD-negative individuals in the Chinese population. Blood Transfus 2014;12:238-43. [PubMed]
- Wang M, Wang BL, Xu W, et al. Anti-D alloimmunisation in pregnant women with DEL phenotype in China. Transfus Med 2015;25:163-9. [Crossref] [PubMed]
- Xu W, Zhu M, Wang BL, et al. Prospective Evaluation of a Transfusion Policy of RhD-Positive Red Blood Cells into DEL Patients in China. Transfus Med Hemother 2015;42:15-21. [Crossref] [PubMed]
- Flegel WA. The above letter was also sent to Dr Flegel: Dr Flegel offered the following reply. Transfusion 2006;46:1063-4. [Crossref] [PubMed]
- Choi S, Chun S, Seo JY, et al. Planned Transfusion of D-Positive Blood Components in an Asia Type DEL Patient: Proposed Modification of the Korean National Guidelines for Blood Transfusion. Ann Lab Med 2019;39:102-4. [Crossref] [PubMed]
- Chun S, Kim H, Yun JW, et al. RHD genotyping is recommended for all patients with serological weak-D phenotypes in Asian populations - Cases with coexistence of weak-D and Asia type DEL alleles results in complete expression of D-antigen. Transfus Apher Sci 2020;59:102807. [Crossref] [PubMed]
- Yan L, Wu J, Zhu F, et al. Molecular basis of D variants in Chinese persons. Transfusion 2007;47:471-7. [Crossref] [PubMed]
- Wagner FF, Frohmajer A, Ladewig B, et al. Weak D alleles express distinct phenotypes. Blood 2000;95:2699-708. [Crossref] [PubMed]
- Mouro I, Le Van Kim C, Rouillac C, et al. Rearrangements of the blood group RhD gene associated with the DVI category phenotype. Blood 1994;83:1129-35. [Crossref] [PubMed]
- Wagner FF, Gassner C, Muller TH, et al. Three molecular structures cause rhesus D category VI phenotypes with distinct immunohematologic features. Blood 1998;91:2157-68. [Crossref] [PubMed]
- Körmöczi GF, Gassner C, Shao CP, et al. A comprehensive analysis of DEL types: partial DEL individuals are prone to anti-D alloimmunization. Transfusion 2005;45:1561-7. [Crossref] [PubMed]
- Borgmann DM, Mayr S, Polin H, et al. Single Molecule Fluorescence Microscopy and Machine Learning for Rhesus D Antigen Classification. Sci Rep 2016;6:32317. [Crossref] [PubMed]
- Subramaniyan R. Prevalence of D variants in the Indian donor population. Hematol Transfus Cell Ther 2019;41:190-3. [Crossref] [PubMed]
- Gu J, Sun AY, Wang XD, et al. Analysis of density and epitopes of D antigen on the surface of erythrocytes from DEL phenotypic individuals carrying the RHD1227A allele. Blood Transfus 2014;12:244-9. [PubMed]
- Zhang X, Li G, Zhou Z, et al. Molecular and computational analysis of 45 samples with a serologic weak D phenotype detected among 132,479 blood donors in northeast China. J Transl Med 2019;17:393. [Crossref] [PubMed]
- Ye L, Li M, Yang Q, et al. RHD alleles contributing to serologically weak D phenotypes in China: A single-centre study over 10 years. Vox Sang 2022;117:949-57. [Crossref] [PubMed]
- Wagner FF, Gassner C, Müller TH, et al. Molecular basis of weak D phenotypes. Blood 1999;93:385-93. [Crossref] [PubMed]
- Silvy M, Chapel-Fernandes S, Callebaut I, et al. Characterization of novel RHD alleles: relationship between phenotype, genotype, and trimeric architecture. Transfusion 2012;52:2020-9. [Crossref] [PubMed]
- Li Q, Ye LY, Guo ZH, et al. Study on the molecular background of Del phenotype in Chinese population. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2006;23:486-91. [PubMed]
- Flegel WA, Wagner FF. RHD epitope density profiles of RHD variant red cells analyzed by flow cytometry. Transfus Clin Biol 1996;3:429-31. [Crossref] [PubMed]
- Krog GR, Clausen FB, Berkowicz A, et al. Is current serologic RhD typing of blood donors sufficient for avoiding immunization of recipients? Transfusion 2011;51:2278-85. [Crossref] [PubMed]
- Flegel WA, von Zabern I, Wagner FF. Six years' experience performing RHD genotyping to confirm D- red blood cell units in Germany for preventing anti-D immunizations. Transfusion 2009;49:465-71. [Crossref] [PubMed]
- Gu J, Wang XD, Shao CP, et al. Molecular basis of DEL phenotype in the Chinese population. BMC Med Genet 2014;15:54. [Crossref] [PubMed]
- Okuda H, Kawano M, Iwamoto S, et al. The RHD gene is highly detectable in RhD-negative Japanese donors. J Clin Invest 1997;100:373-9. [Crossref] [PubMed]
- Leger RM, Arndt PA, Ciesielski DJ, et al. False-positive eluate reactivity due to the low-ionic wash solution used with commercial acid-elution kits. Transfusion 1998;38:565-72. [Crossref] [PubMed]
- Kim TY, Hong YJ, Kim MJ, et al. Recommendations Regarding Practical DEL Typing Strategies for Serologically D-Negative Asian Donors. Transfus Med Hemother 2020;47:88-93. [Crossref] [PubMed]
- Richard M, Perreault J, Constanzo-Yanez J, et al. A new DEL variant caused by exon 8 deletion. Transfusion 2007;47:852-7. [Crossref] [PubMed]
- Pisacka M, Kralova J, Hanzikova V, et al. Strong Pregnancy induced anti-D immunization in Del phenotype with RHD*01EL.04 allele. Abstract 5A-229-04. Vox Sang 2019;114:1.
- Singleton BK, Green CA, Kumura K, et al. Two new RHD mutations associated with the DEL phenotype. Transf Clin Biol 2001;8:1.
- Christiansen M, Sørensen BS, Grunnet N. RHD positive among C/E+ and D- blood donors in Denmark. Transfusion 2010;50:1460-4. [Crossref] [PubMed]
- Gassner C, Doescher A, Drnovsek TD, et al. Presence of RHD in serologically D-, C/E+ individuals: a European multicenter study. Transfusion 2005;45:527-38. [Crossref] [PubMed]
- Ye L, He Y, Gao H, et al. Weak D phenotypes caused by intronic mutations in the RHD gene: four novel weak D alleles identified in the Chinese population. Transfusion 2013;53:1829-33. [Crossref] [PubMed]
- Wagner FF, Mardt I, Bittner R, et al. RHD PCR of blood donors in Northern Germany: Use of adsorption/elution to determine D antigen status. Vox Sang 2012;103:1.
- Stegmann TC, Veldhuisen B, Bijman R, et al. Frequency and characterization of known and novel RHD variant alleles in 37 782 Dutch D-negative pregnant women. Br J Haematol 2016;173:469-79. [Crossref] [PubMed]
- Müller SP, Bartels I, Stein W, et al. The determination of the fetal D status from maternal plasma for decision making on Rh prophylaxis is feasible. Transfusion 2008;48:2292-301. [Crossref] [PubMed]
- Silvy M, Simon S, Gouvitsos J, et al. Weak D and DEL alleles detected by routine SNaPshot genotyping: identification of four novel RHD alleles. Transfusion 2011;51:401-11. [Crossref] [PubMed]
- Le Maréchal C, Guerry C, Benech C, et al. Identification of 12 novel RHD alleles in western France by denaturing high-performance liquid chromatography analysis. Transfusion 2007;47:858-63. [PubMed]
- Raud L, Le Tertre M, Vigneron L, et al. Missense RHD single nucleotide variants induce weakened D antigen expression by altering splicing and/or protein expression. Transfusion 2021;61:2468-76. [Crossref] [PubMed]
- Garcia F, Rodriguez MA, Goldman M, et al. New RHD variant alleles. Transfusion 2015;55:427-9. [Crossref] [PubMed]
- Trucco Boggione C, Nogués N, González-Santesteban C, et al. Characterization of RHD locus polymorphism in D negative and D variant donors from Northwestern Argentina. Transfusion 2019;59:3236-42. [Crossref] [PubMed]
- Turley E, McGowan EC, Hyland CA, et al. Severe hemolytic disease of the fetus and newborn due to allo-anti-D in a patient with a partial DEL phenotype arising from the variant allele described as RHD*148+1T (RHD*01EL.31). Transfusion 2018;58:2260-4. [Crossref] [PubMed]
- Kim TY, Yu H, Cho D. The intronic variant RHD:c.149-29G>C designated as RHD*01EL.32 does not cause a DEL phenotype. Transfusion 2021;61:986-7. [Crossref] [PubMed]
- Tounsi WA, Madgett TE, Avent ND. Complete RHD next-generation sequencing: establishment of reference RHD alleles. Blood Adv 2018;2:2713-23. Erratum in: Blood Adv 2019;3:120. [Crossref] [PubMed]
- Orzińska A, Guz K, Polin H, et al. RHD variants in Polish blood donors routinely typed as D-. Transfusion 2013;53:2945-53. [PubMed]
- von Zabern I, Flegel WA. IVS5-38del4 deletion in the RHD gene does not cause a DEL phenotype: relevance for RHD alleles including DFR-3. Transfusion 2007;47:1552-5. [Crossref] [PubMed]
- Wagner F, Doscher A, Mardt I. Results of more than ten years testing of RhD negative first time donors by RHD PCR. Transfus Med Hemother 2019;46:1.
- Crottet SL, Henny C, Meyer S, et al. Implementation of a mandatory donor RHD screening in Switzerland. Transfus Apher Sci 2014;50:169-74. [Crossref] [PubMed]
- Trucco Boggione C, Luján Brajovich ME, Tarragó M, et al. Molecular structures identified in serologically D- samples of an admixed population. Transfusion 2014;54:2456-62. [Crossref] [PubMed]
- Pham BN, Roussel M, Gien D, et al. Molecular analysis of patients with weak D and serologic analysis of those with anti-D (excluding type 1 and type 2). Immunohematology 2013;29:55-62. [Crossref] [PubMed]
- Isa K, Sasaki K, Ogasawara K, et al. Prevalence of RHD alleles in Japanese individuals with weak D phenotype: Identification of 20 new RHD alleles. Vox Sang 2016;111:315-9. [Crossref] [PubMed]
- Lopez GH, McGowan EC, Condon JA, et al. Genotyping by sequencing defines independent novel RHD variants for an antenatal patient and a blood donor. Transfusion 2017;57:2281-3. [Crossref] [PubMed]
- Lopez GH, Turner RM, McGowan EC, et al. A DEL phenotype attributed to RHD Exon 9 sequence deletion: slipped-strand mispairing and blood group polymorphisms. Transfusion 2018;58:685-91. [Crossref] [PubMed]
- Chun S, Lee HJ, Cho D. A new RHD variant allele (RHD Gly339Val) shows weakened D expression compared to RHD Gly339Glu and Gly339Arg mutants. Transfusion 2020;60:E17-8. [Crossref] [PubMed]
- de Paula Vendrame TA, Prisco Arnoni C, Guilhem Muniz J, et al. Characterization of RHD alleles present in serologically RHD-negative donors determined by a sensitive microplate technique. Vox Sang 2019;114:869-75. [Crossref] [PubMed]
- Stettler J, Lejon Crottet S, Still F, et al. Reduced RhD antigen expression caused by two novel RHD variant alleles. Transfusion 2020;60:E7-8. [Crossref] [PubMed]
- Polin H, Danzer M, Hofer K, et al. Effective molecular RHD typing strategy for blood donations. Transfusion 2007;47:1350-5. [Crossref] [PubMed]
- Faas BH, Beckers EA, Simsek S, et al. Involvement of Ser103 of the Rh polypeptides in G epitope formation. Transfusion 1996;36:506-11. [Crossref] [PubMed]
- Srivastava K, Stiles DA, Wagner FF, et al. Two large deletions extending beyond either end of the RHD gene and their red cell phenotypes. J Hum Genet 2018;63:27-35. [Crossref] [PubMed]
- El Housse H, El Wafi M, Ouabdelmoumene Z, et al. Comprehensive phenotypic and molecular investigation of RhD and RhCE variants in Moroccan blood donors. Blood Transfus 2019;17:151-6. [PubMed]
- Chang JG, Wang JC, Yang TY, et al. Human RhDel is caused by a deletion of 1,013 bp between introns 8 and 9 including exon 9 of RHD gene. Blood 1998;92:2602-4. [Crossref] [PubMed]
- Gowland P, Gassner C, Hustinx H, et al. Molecular RHD screening of RhD negative donors can replace standard serological testing for RhD negative donors. Transfus Apher Sci 2014;50:163-8. [Crossref] [PubMed]
- Polin H, Danzer M, Gaszner W, et al. Identification of RHD alleles with the potential of anti-D immunization among seemingly D- blood donors in Upper Austria. Transfusion 2009;49:676-81. [Crossref] [PubMed]
- Fichou Y, Le Maréchal C, Bryckaert L, et al. Variant screening of the RHD gene in a large cohort of subjects with D phenotype ambiguity: report of 17 novel rare alleles. Transfusion 2012;52:759-64. [Crossref] [PubMed]
- Fichou Y, Chen JM, Le Maréchal C, et al. Weak D caused by a founder deletion in the RHD gene. Transfusion 2012;52:2348-55. [Crossref] [PubMed]
- Just B, Deitenbeck R. Sekundäre Anti-D-Immunisierung bei RhD-negativem Empfänger, verursacht durch ein Del-Erythrozytenkonzentrat. Transfusionsmedizin - Immunhämatologie, Hämotherapie, Immungenetik, Zelltherapie. 2014;4:134-8. Available online: https://www.thieme-connect.com/products/ejournals/abstract/10.1055/s-0034-1368630
- Esteban R, Montero R, Flegel WA, et al. The D category VI type 4 allele is prevalent in the Spanish population. Transfusion 2006;46:616-23. [Crossref] [PubMed]
- Safic Stanic H, Dogic V, Herceg I, et al. D variants in the population of D-negative blood donors in the north-eastern region of Croatia. Transfus Med 2021;31:43-7. [Crossref] [PubMed]
- den Dunnen JT, Dalgleish R, Maglott DR, et al. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum Mutat 2016;37:564-9. [Crossref] [PubMed]
- Raud L, Ka C, Gourlaouen I, et al. Functional analysis of novel RHD variants: splicing disruption is likely to be a common mechanism of variant D phenotype. Transfusion 2019;59:1367-75. [Crossref] [PubMed]
- Fichou Y, Gehannin P, Corre M, et al. Extensive functional analyses of RHD splice site variants: Insights into the potential role of splicing in the physiology of Rh. Transfusion 2015;55:1432-43. [Crossref] [PubMed]
- Flegel WA, Eicher NI, Doescher A, et al. In-frame triplet deletions in RHD alter the D antigen phenotype. Transfusion 2006;46:2156-61. [Crossref] [PubMed]
- Ogasawara K, Sasaki K, Isa K, et al. Weak D alleles in Japanese: a c.960G>A silent mutation in exon 7 of the RHD gene that affects D expression. Vox Sang 2016;110:179-84. [Crossref] [PubMed]
- Choi Y, Sims GE, Murphy S, et al. Predicting the functional effect of amino acid substitutions and indels. PLoS One 2012;7:e46688. [Crossref] [PubMed]
- Sim NL, Kumar P, Hu J, et al. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 2012;40:W452-7. [Crossref] [PubMed]
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248-9. [Crossref] [PubMed]
- Floch A, Vege S, Berardi P, et al. An intron c.149-2632T>A change in RHD is associated with aberrant transcription and very weak D phenotype. Transfusion 2022;62:E14-E16. [Crossref] [PubMed]
- Chen DP, Sun CF, Ning HC, et al. Comprehensive analysis of RHD splicing transcripts reveals the molecular basis for the weak anti-D reactivity of Del -red blood cells. Transfus Med 2016;26:123-9. [Crossref] [PubMed]
- Lin JH, Wu H, Zou WB, et al. Splicing Outcomes of 5' Splice Site GT>GC Variants That Generate Wild-Type Transcripts Differ Significantly Between Full-Length and Minigene Splicing Assays. Front Genet 2021;12:701652. [Crossref] [PubMed]
- Rouillac C, Colin Y, Hughes-Jones NC, et al. Transcript analysis of D category phenotypes predicts hybrid Rh D-CE-D proteins associated with alteration of D epitopes. Blood 1995;85:2937-44. [Crossref] [PubMed]
- Flegel WA, Srivastava K. Frameshift variations in the RHD coding sequence: Molecular mechanisms permitting protein expression. Transfusion 2020;60:2737-44. [Crossref] [PubMed]
- Gruswitz F, Chaudhary S, Ho JD, et al. Function of human Rh based on structure of RhCG at 2.1 A. Proc Natl Acad Sci U S A 2010;107:9638-43. [Crossref] [PubMed]
- Conroy MJ, Bullough PA, Merrick M, et al. Modelling the human rhesus proteins: implications for structure and function. Br J Haematol 2005;131:543-51. [Crossref] [PubMed]
- Costa S, Martin F, Chiba A, et al. RHD alleles and D antigen density among serologically D- C+ Brazilian blood donors. Transfus Med 2014;24:60-1. [Crossref] [PubMed]
- Guzijan G, Jovanovic Srzentic S, Pavlovic Jankovic N, et al. Implementation of Molecular RHD Typing at Two Blood Transfusion Institutes from Southeastern Europe. Transfus Med Hemother 2019;46:114-20. [Crossref] [PubMed]
- Credidio DC, Pellegrino J, Castilho L. Serologic and molecular characterization of D variants in Brazilians: impact for typing and transfusion strategy. Immunohematology 2011;27:6-11. [Crossref] [PubMed]
- Cruz BR, Chiba AK, Moritz E, et al. RHD alleles in Brazilian blood donors with weak D or D-negative phenotypes. Transfus Med 2012;22:84-9. [Crossref] [PubMed]
- Mota M, Dezan M, Valgueiro MC, et al. RHD allelic identification among D-Brazilian blood donors as a routine test using pools of DNA. J Clin Lab Anal 2012;26:104-8. [Crossref] [PubMed]
- Dezan MR, Guardalini LGO, Pessoa E, et al. Evaluation of the applicability and effectiveness of a molecular strategy for identifying weak D and DEL phenotype among D- blood donors of mixed origin exhibiting high frequency of RHD*Ψ. Transfusion 2018;58:317-22. [Crossref] [PubMed]
- St-Louis M, Lebrun A, Goldman M, et al. Alloimmunization of patients by blood units harboring distinct DEL variants. Immunohematology 2013;29:136-40. [Crossref] [PubMed]
- Xu Q, Grootkerk-Tax MG, Maaskant-van Wijk PA, et al. Systemic analysis and zygosity determination of the RHD gene in a D-negative Chinese Han population reveals a novel D-negative RHD gene. Vox Sang 2005;88:35-40. [Crossref] [PubMed]
- Li Q, Ye LY, Guo ZH, et al. Molecular basis of D variants between Uigur and Han blood donors in Xinjiang. Transfus Med 2008;18:199-203. [Crossref] [PubMed]
- Chen Q, Li M, Li M, et al. Molecular basis of weak D and DEL in Han population in Anhui Province, China. Chin Med J (Engl) 2012;125:3251-5. [PubMed]
- Wu X, Wu D, Wang M, et al. Analysis of frequency of a RHD1227A allele in Chinese Hans. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2014;31:793-6. [PubMed]
- Peng CT, Shih MC, Liu TC, et al. Molecular basis for the RhD negative phenotype in Chinese. Int J Mol Med 2003;11:515-21. [Crossref] [PubMed]
- Lee YL, Chiou HL, Hu SN, et al. Analysis of RHD genes in Taiwanese RhD-negative donors by the multiplex PCR method. J Clin Lab Anal 2003;17:80-4. [Crossref] [PubMed]
- Dajak S, Krstic JL, Körmöczi G, et al. Characteristics and frequency of DEL phenotype detected by indirect antiglobulin test in Dalmatia county of Croatia. Transfus Apher Sci 2014;50:210-3. [Crossref] [PubMed]
- Tammi SM, Tounsi WA, Sainio S, et al. Next-generation sequencing of 35 RHD variants in 16 253 serologically D- pregnant women in the Finnish population. Blood Adv 2020;4:4994-5001. [Crossref] [PubMed]
- Samir S, Jain A, Marwaha N. Frequency of DEL phenotype in RhD negative donor population of north India. Transfus Apher Sci 2015;53:34-7. [Crossref] [PubMed]
- Kulkarni S, Parchure DS, Gopalkrishnan V, et al. Screening for DEL phenotype in RhD negative Indians. J Clin Lab Anal 2018;32:e22288. [Crossref] [PubMed]
- Chaudhary R, Verma S, Verma A. Prevalence of DEL phenotype in D- blood donors in India. Immunohematology 2020;36:133-6. [Crossref] [PubMed]
- Londero D, Fiorino M, Miotti V, et al. Molecular RH blood group typing of serologically D-/CE+ donors: the use of a polymerase chain reaction-sequence-specific primer test kit with pooled samples. Immunohematology 2011;27:25-8. [Crossref] [PubMed]
- Seo MH, Won EJ, Hong YJ, et al. An effective diagnostic strategy for accurate detection of RhD variants including Asian DEL type in apparently RhD-negative blood donors in Korea. Vox Sang 2016;111:425-30. [Crossref] [PubMed]
- Kim B, Lee ST, Kim S, et al. Application of Multiplex Ligation-Dependent Probe Amplification Assay for Genotyping Major Blood Group Systems Including DEL Variants in the D-Negative Korean Population. Ann Lab Med 2018;38:32-8. [Crossref] [PubMed]
- Chung YN, Kim TY, Yu H, et al. Molecular basis of serological weak D phenotypes and RhD typing discrepancies identified in the Korean population. Blood Transfus 2021;19:327-34. [PubMed]
- Kabiri Z, Benajiba M, Hajjout K, et al. Serological analysis of phenotype Del among blood donors Rh D negative Moroccan. Pathol Biol (Paris) 2015;63:111-2. [Crossref] [PubMed]
- Wah ST, Chi SN, Kyaing KK, et al. Serological Detection of Rh-Del Phenotype among Rh-Negative Blood Donors at National Blood Center, Yangon, Myanmar. Adv Hematol 2020;2020:3482124. [Crossref] [PubMed]
- Moussa H, Tsochandaridis M, Chakroun T, et al. Molecular background of D-negative phenotype in the Tunisian population. Transfus Med 2012;22:192-8. [Crossref] [PubMed]
- Liu J, Zhang S, Wang Q, et al. Frequencies and ethnic distribution of ABO and RhD blood groups in China: a population-based cross-sectional study. BMJ Open 2017;7:e018476. [Crossref] [PubMed]
- Fujita Y, Tanaka K, Tanimura M. The distribution of the Rh(D) blood types in Japan. Jinrui Idengaku Zasshi 1978;23:197-209. [Crossref] [PubMed]
- Polin H, Gaszner W, Hackl C, et al. On the trail of anti-CDE to unexpected highlights of the RHD*weak 4.3 allele in the Upper Austrian population. Vox Sang 2012;103:130-6. [Crossref] [PubMed]
- Gardener GJ, Legler TJ, Hyett JA, et al. Anti-D in pregnant women with the RHD(IVS3+1G>A)-associated DEL phenotype. Transfusion 2012;52:2016-9. [Crossref] [PubMed]
- Yasuda H, Ohto H, Sakuma S, et al. Secondary anti-D immunization by Del red blood cells. Transfusion 2005;45:1581-4. [Crossref] [PubMed]
- Kim KH, Kim KE, Woo KS, et al. Primary anti-D immunization by DEL red blood cells. Korean J Lab Med 2009;29:361-5. [PubMed]
- Shao CP, Wang BY, Ye SH, et al. DEL RBC transfusion should be avoided in particular blood recipient in East Asia due to allosensitization and ineffectiveness. J Zhejiang Univ Sci B 2012;13:913-8. [Crossref] [PubMed]
- Yang HS, Lee MY, Park TS, et al. Primary anti-D alloimmunization induced by "Asian type" RHD (c.1227G>A) DEL red cell transfusion. Ann Lab Med 2015;35:554-6. [Crossref] [PubMed]
- Yoon J, Koh YE, Kim HN, et al. A Case of Primary Anti-D Alloimmunization by RHD(c.1227G>A) DEL Red Blood Cell Transfusion. The Korean Journal of Blood Transfusion. 2016;27:169-73. [Crossref]
- Chen W, Li L, Tsai SL. Anti-D immunization by Del red blood cells in Taiwan: two case reports. Transfusion 2006;46:1.
- Safic Stanic H, Dogic V, Bingulac-Popovic J, et al. RhD alloimmunization by DEL variant missed in donor testing. Transfusion 2022;62:1084-8. [Crossref] [PubMed]
- Ji Y, Luo G, Fu Y. Incidence of anti-D alloimmunization in D-negative individuals receiving D-positive red blood cell transfusion: A systematic review and meta-analysis. Vox Sang 2022;117:633-40. [Crossref] [PubMed]
- Flegel WA, Wagner FF, Ó Donghaile DP. Anti-D immunization rates may exceed 50% in many clinically relevant settings, despite varying widely among patient cohorts. Transfusion 2020;60:1109-10. [Crossref] [PubMed]
- Flommersfeld S, Mand C, Kühne CA, et al. Unmatched Type O RhD+ Red Blood Cells in Multiple Injured Patients. Transfus Med Hemother 2018;45:158-61. [Crossref] [PubMed]
- Selleng K, Jenichen G, Denker K, et al. Emergency transfusion of patients with unknown blood type with blood group O Rhesus D positive red blood cell concentrates: a prospective, single-centre, observational study. Lancet Haematol 2017;4:e218-24. [Crossref] [PubMed]
- Reid ME. Transfusion in the age of molecular diagnostics. Hematology Am Soc Hematol Educ Program 2009;171-7. [Crossref] [PubMed]
- Schmidt PJ. Are weak D red blood cells really immunogenic? Transfusion 2006;46:2029-30. [Crossref] [PubMed]
- Sainio S, Nupponen I, Kuosmanen M, et al. Diagnosis and treatment of severe hemolytic disease of the fetus and newborn: a 10-year nationwide retrospective study. Acta Obstet Gynecol Scand 2015;94:383-90. [Crossref] [PubMed]
- de Haas M, Thurik FF, Koelewijn JM, et al. Haemolytic disease of the fetus and newborn. Vox Sang 2015;109:99-113. [Crossref] [PubMed]
- Bundesärztekammer, Richtlinie zur Gewinnung von Blut und Blutbestandteilen und zur Anwendung von Blutprodukten (Richtlinie Hämotherapie), Gesamtnovelle 2017, umschriebene Fortschreibung 2021, (2021). Available online: https://www.bundesaerztekammer.de/fileadmin/user_upload/_old-files/downloads/pdf-Ordner/RL/RiliH_Lese.pdf
Cite this article as: Wagner FF. Serology and molecular biology of DEL: a narrative review. Ann Blood 2023;8:28.


