
The power of digital PCR in fetal blood group genotyping: a review
Introduction and aim
Early assessment of the fetal blood group status provides useful clinical information to guide the perinatal care for pregnant women who have been alloimmunized to a clinically significant red cell antigen. Should the fetus inherit the paternal red cell antigen targeted by the maternal antibody there is a risk of the fetus developing haemolytic disease of the fetus and newborn (HDFN). In such cases, expert antenatal monitoring is indicated to detect and, if indicated, respond to any signs of significant fetal anaemia. Delivery planning and newborn care may also be altered accordingly. Conversely, if the fetus has not inherited the paternal antigen of interest, expert antenatal monitoring, earlier delivery, tertiary newborn care and unnecessary maternal anxiety can all be avoided.
Liquid biopsy, a non-invasive alternative to chorionic villus sampling (CVS) or amniocentesis, permits non-invasive prenatal testing (NIPT) of circulating cell-free fetal (cff)DNA in the maternal plasma, enabling detection of paternally inherited markers (1). NIPT is based on discoveries led by Professor Lo et al. from late last century, that placental derived cffDNA circulates in the maternal plasma against a background of maternal cell-free deoxyribonucleic acid (DNA), defined as mcfDNA (2,3). The concentration of cffDNA increases with gestation and is cleared rapidly after delivery, thereby representing fetal markers specific for that pregnancy. There are three characteristics of cffDNA that pose challenges with NIPT: namely the cffDNA comprises smaller size fragments than mcfDNA (4); cffDNA is present in a low concentration and is present in the maternal circulation as a low abundant molecule against a background of more abundant and larger fragments of mcfDNA (2).
Fetal blood group genotyping to predict the RhD antigen status served as one of the first models to demonstrate the capability and clinical utility for NIPT technology (2,5). RhD-negative women, particularly with a Caucasian background, are most commonly homozygous for the RHD*01N.01 haplotype, in which the RHD gene is deleted at the RH blood group locus (6). As such, there is no competing background maternal RHD cfDNA in the circulation and NIPT for fetal RHD can be performed using real-time or quantitative PCR, defined as qPCR, instrumentation. Fetal blood group RHD genotyping was therefore amongst the earliest clinical applications for NIPT technology and was applied as a diagnostic for alloimmunized women, presenting with allo-anti-D (5,7,8).
After RhD, the most clinically significant red cell antigens are Kell (K), Rhc and RhE and the Duffy antigens, Fya and Fyb. It has been noted that mothers alloimmunized with anti-K and anti-c, in particular, are at high risk for having a fetus affected by HDFN (>50% risk) if the infant inherits the respective paternal antigen (9). However, the application of NIPT in such cases is more problematic as these, and indeed many other blood group antigens arise, from single nucleotide variants (SNVs) on the blood group gene. This poses a problem using qPCR techniques, as the allele from the mcfDNA is more abundant and will compete with the allele from the cffDNA in the PCR reaction (10-12).
The aim of this paper is to discuss the power for digital PCR, defined as dPCR, to overcome the limitations imposed by the intrinsic characteristics of cffDNA and to discuss the potential for dPCR to serve as an additional clinical diagnostic tool for fetal blood group genotyping. The scope is limited to application of dPCR for blood groups and the reader is referred to other reviews for a broader application of NIPT dPCR for detection of fetal chromosomal anomalies and single gene disorders.
Principle behind digital (d) PCR
The digital (d) PCR is based on first diluting and compartmentalizing individual DNA molecules into individual PCR mixes, so that the competing (maternal) molecules are separated from the target (fetal) DNA molecule prior to amplification (13). Digital (d) PCR in essence dilutes an initial PCR reaction mixture into thousands to tens of thousands of reaction mixtures such that each mixture comprises 0 or 1 copy of a DNA template from the initial mixture. The majority of reaction mixtures comprise zero copies and the remaining contain either maternal or fetal cfDNA partitioned away from each other. Given the maternal and fetal DNA differ in the target gene by a SNV, this separation means that the probe, specific for the paternally inherited SNV on fetal DNA, will not be affected by the excessive abundance of maternal DNA in the PCR reaction.
Different dPCR platforms have been developed and include droplet digital PCR, defined as ddPCR, where PCR reactions are performed under oil emulsion, and Microfluidics PCR where reactions occur in DNA microchip chambers. For a recent review the reader is referred to Nogués, 2020 (14) as well as to Hindson et al., 2011 and Quan et al., 2018 for further background and pictorial depictions (15,16). Independent of the platform, each PCR reaction for the diluted mixture runs to an end point, after which fluorescent signal is scored. For example, with ddPCR, the droplet reader sips the reaction mixture and singulates the droplets to read fluorescent signals from each reaction to score the number of positive or negative fluorescence signals. The software calculates the fraction of positive and negative droplets and uses Poisson statistics to calculate the concentration of the target molecule. Should the assay design include the alternate allele rather than just the target allele, the software will also calculate the fractional abundance of rare sequences.
Feasibility studies for dPCR
The feasibility for dPCR was first shown by Vogelstein and Kinzler in 1999 to address the problem in cancer research on how to detect a small number of tumour derived alleles among a large excess of normal cells (17). DNA from colorectal cancer cell lines was diluted and subsequent dPCR products were derived from single molecule templates. Sequencing showed that the PCR products were either completely variant or completely wild-type. They concluded that “Dig-PCR” (now dPCR) can be used to detect mutations present at relatively low levels in the samples to be analysed. The limit of detection was defined by the number of wells that can be analyzed and the intrinsic mutation rate of the polymerase used for amplification. As the name implies, dPCR is quantitative and counts the number of positive signals which should be distributed according to Poisson probabilities (17).
In 2007, Fan and Quake predicted that the dPCR method may be applicable to NIPT to detect the presence of fetal markers testing the cffDNA (18). Their prediction was predicated on the experiment at the time showing dPCR accurately detected trisomy 21 (down syndrome) in mixtures of human genomic DNA from a normal cell line and a trisomy 21 cell line (18). They therefore foresaw the practical application of applying dPCR to detect fetal cfDNA, which, as in their in-vitro study, is mixed with maternal cfDNA.
Subsequently, Lun et al. [2008] tested the hypothesis that microfluidics dPCR would enhance the precision of measuring circulating fetal DNA (19). Using an artificial sample mixture of 7% male DNA they carried out 20 analyses on the dPCR platform and non-digital PCR. The CVs of the digital and nondigital assays were 16% and 49%, indicating that the digital assay was 3.1 times more precise than the nondigital assay. The study also recruited 10 pregnant women with male fetuses for each trimester and 5 pregnant women with female fetuses in the first trimester for the comparison of three PCR platforms: dPCR, qPCR and mass spectrometry using PCR assays designed to target the ZFY (zinc finger protein, Y-linked) gene. The dPCR showed improved sensitivity—100% versus 90%—in the pregnancies with male fetuses. Also of note, the median fraction of fetal-DNA concentration was higher with dPCR than qPCR with fractional fetal-DNA measured as 9.7%, 9.0% and 20.4% for the first second and third trimesters by dPCR compared to 4.8%, 4.1% and 7.6% reported by qPCR. This paper became a reference for acknowledging that cffDNA levels present in maternal plasma were more abundant than first thought.
Clinical feasibility for NIPT dPCR for fetal blood group SNV genotyping
The first study into dPCR for fetal blood group assessment involved a collaborative study between Professor Lo’s team in Hong Kong and Australian Red Cross Lifeblood researchers. This involved a retrospective study involving two women who demonstrated a rare SNV on the RHD gene at an exon/intron boundary, defined as RHD*01DEL8 (previously known as RHD*IVS3+1G>A) (13). This RHD variant has a low prevalence in European populations (1 per 15,152 of the population) and is associated with a very weak RhD phenotype, called DEL (20). Red cells with this phenotype express a quantitatively reduced antigen density, detected by adsorbing and eluting anti-D antibodies from the red cell. This (and some of the other DEL phenotypes) also exhibits a qualitative difference resulting in partial D antigen expression, such that mothers can make forms of anti-D antibody directed against the missing D-epitopes (21-24). Such anti-D have been associated with HDFN where the baby is RhD positive (22). Tsui et al. showed the feasibility for a microfluidic dPCR approach to determine whether the fetal-derived wild type RHD allele sequences could be detected against a background of the dominant maternal RHD SNV-based variant (13).
The approach by Tsui et al., first showed the dPCR specificity and sensitivity using genomic DNA mixtures to mimic fractions of fetal cfDNA relative to the maternal cfDNA of between 2% to 10% for this RHD variant genotyping scenario. They then tested two cases. For Case 1 with a male fetus, sampled at 34 weeks, the fetal fraction (ff) assessed from informative SNV markers was 8.20%: the dPCR used a 4,590 well format and yielded positive signals among 23 of the 4,590 wells, predicting an RhD-positive fetus. For Case 2 with a female fetus, sampled at 15 weeks, the fetal fraction was 4.15%: the dPCR yielded positive signals in 11 wells among the 11,475 wells tested. This resulted in the prediction of an RhD-positive fetus (13). For both cases, the predictions were confirmed using the cord blood RhD phenotype.
The two cases illustrate the power of the system, partitioning (or subdividing) nucleic acid samples into thousands of nanoliter-sized droplets. This is combined with the capability of scoring fluorescent signals, using automated processes within dPCR systems. Here both cases were clinically significant and this RHD variant represents one of many blood group allelic variants arising from SNVs that are associated with a clinically significant phenotype.
Comparison of dPCR to qPCR for mothers with the deleted RHD haplotype
Before considering NIPT dPCR for other blood group SNVs it is worth reviewing two studies that compared the performance of ddPCR against the established qPCR for fetal RHD genotyping where the RhD-negative mother has the deleted RHD haplotype (RHD*01N/01N).
Sillence et al. [2015] enrolled 46 RhD negative pregnant women with the deleted RHD*01N.01 haplotype (10). Maternal blood samples were collected in either Streck cell-free DNA BCT (n=24) or ethylenediaminetetraacetic acid (EDTA) (n=22) tubes, the former designed to stabilize white cells and minimize lysis. By ddPCR, genotyping for fetal RHD exons 5 and 7 was accurate in 100% (24/24) of Streck samples and 95.5% (21/22) of EDTA samples. In contrast to these ddPCR outcomes, qPCR detected RHD exon 5 with a reduced accuracy of 83% in samples with optimal fetal cfDNA fractions (>4%). The accuracy of qPCR compared to dPCR was reduced further, for RHD exon 5 and 7, when the fetal fraction had been suboptimal (<2%). They concluded false qPCR results were not caused by low cffDNA concentrations, since these were similar to ‘optimal’ samples, but by low concentration of cffDNA relative to high maternal cfDNA i.e., low fetal fraction, attributed to possible competition with the homologous maternal RHCE allele. They noted overall qPCR was considered more susceptible to non-specific amplification of the maternal allele and dPCR more powerful in the detection of alleles associated with conditions associated with a SNV (giving B-thalassemia and cystic fibrosis as examples) (10).
Svobodová et al. [2015] compared ddPCR with qPCR using diluted genomic DNA of known concentrations. The PCR assay included assaying three RHD exons (5, 7 and 10). In their study, dPCR displayed lower variability between replicates. The detection limits in their study and the linearity of the assays were comparable (limits near 1 Genome Equivalent). In a second stage, clinical plasma samples were analysed for 35 RhD-negative pregnant women, with 25 of these women carrying an RHD positive fetus. The fetal fraction measurements were higher for ddPCR compared with qPCR (15.7% versus 9.8%) and correlated with gestational age. It was noted that with ddPCR that the operator has to manually set the fluorescence threshold levels which defines the cut-off for interpreting the number of fluorescence signals as positive or negative (25).
Diversity of SNV based blood group antigens and clinical significance
The International Society of Blood Transfusion (ISBT) Working Party for Red Cell Immunogenetics and Blood Group Terminology currently recognizes 43 blood group systems, each with a defined but separate locus across the human genome (26). Within these systems, 345 antigens have been characterized, many of them defined by investigations of pregnant women presenting with an unidentified antibody to a red cell antigen (27). Maternal red cell alloantibodies of clinical significance vary among different ethnicities (28,29). A study of 66,354 pregnant women in Australia found that 0.73% harboured antibodies to red cell antigens with at least 20 different antibodies detected and classified into three groups according to clinical significance (30). The Rh antibodies, anti-E and anti-D, were shown to represent 27.6% and 10.4% of all antibodies respectively, after which the most prevalent and significant were anti-K (9.1%) in the Kell system and anti-c (8.7%) in the Rh system and anti-Fya or anti-Fyb (3.1%) in the Duffy blood group system.
Clinical feasibility for ddPCR assay for fetal KEL, Duffy and Rh typing
Based on red cell antibody prevalence and significance in the Australian context, O’Brien et al. developed a suite of ddPCR NIPT assays for K/k, Fya/Fyb, C/c and E fetal typing for pregnant women with the corresponding antibody (11). Initial reporting of 43 out of 81 referred clinical cases showed all ddPCR NIPT genotyping predictions were concordant with the infant blood group phenotype, where provided, resulting in an accuracy of 100% [confidence interval (CI): 92.0–100.0%]. It is worth noting the lower CI level is a measure of the sample size in this ongoing clinical validation study (11).
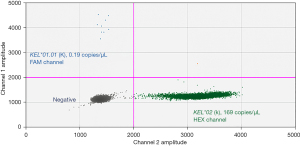
Key considerations for clinical application are the pre-analytical requirements for patient enrolment and for sample processing (time between collection and separation of plasma). The study by O’Brien et al., 2020, shows that fetal K signals can be detected as early as 10 and 12 weeks gestation (correlating with the infant cord phenotype) (Figure 1). From among samples received, the average transport time was 57 hours and fetal blood group genotyping outcomes were concordant for samples collected in either EDTA blood collection tubes or tubes designed to minimize white cell lysis by incorporating a stabilizer. This latter tube type minimizes contamination of the plasma from maternal genomic DNA arising from white cell lysis during transport of the blood sample.
Gaps and limitations for dPCR
There is a need for economic modeling of the relative cost and benefit between qPCR and dPCR technologies. Interestingly there are few studies comparing the accuracy between qPCR and dPCR for detecting fetal blood group genes based on SNV variations. D’Aversa et al., 2018, showed that ddPCR was reliable for detecting the SRY gene marker among 29 samples collected between 4.5 and 12 weeks gestation, noting only 8 could be assigned by qPCR in this early gestation age (31). Table 1 summarizes three further studies which compare qPCR and dPCR for samples between 12 and 39 weeks gestation. These studies were testing for male linked markers or RHD exons rather than SNVs.
Table 1
| Study | Gene target | Sample number | Collection tube | Accuracy (%) | Gestation | |
|---|---|---|---|---|---|---|
| qPCR | dPCR | |||||
| Lun et al. 2008, (19) | ZFX, ZFY | 10 | EDTA | 90% | 100% | 12–39 |
| Sillence et al. 2015, (10) | RHD; RHD exons 5, 7 | 22 | EDTA | 41% | 96% | 28–30 |
| 24 | Cell-Stabilizer Tube with EDTA | 100% (RHD exon 7); 83% (RHD exon 5) | 100% | |||
| Svobodová et al. 2015, (25) | RHD; RHD exons 5, 7, 10 | 35 | EDTA | 100% | 100% | 12–36 |
qPCR, quantitative polymerase chain reaction; dPCR, digital polymerase chain reaction; EDTA, ethylenediaminetetraacetic acid.
NIPT for fetal blood group antigen prediction is most valuable when the genotyping assay used is personalised according to the maternal antibodies that present during the pregnancy. The clinical significance for red cell antibodies varies such that the associated HDFN ranges from mild to very severe with high morbidity and mortality (32). Evidence suggests women with multiple red cell antibodies are more likely to develop significant HDFN than those with a single antibody as reported in 13% of alloimmunized cases in one study (33,34). One of the limitations of the ddPCR approach is its low assay multiplexing capability which means that separate assays need to be established and validated for each blood group antigen. Where multiple antibody specificities are identified in a pregnancy, more reagents, cfDNA template (and consequently more maternal plasma sample) are required to complete the requested investigations.
There is also a need to incorporate robust and universal internal control markers for cases where the fetus types negative for a target blood group allele because in such cases the question arises as to whether cffDNA was present and or extracted correctly from the starting plasma sample. One safeguard to manage this problem is to request a second sample at a subsequent gestation time to confirm and verify false outcomes do not arise from low cffDNA levels in the initial sample.
There is also a recognised problem with receiving sufficient samples for any one clinical marker to validate the test. This is further compounded by the logistical difficulties in obtaining either the cord blood group or infant samples following birth. This is in contrast to validating NIPT fetal RHD genotyping assays since infant cord RhD phenotyping is an established part of antenatal care to target postnatal anti-D prophylaxis for RhD-negative women.
The future for dPCR in combination with MPS technologies
Digital PCR was initially developed in parallel with next-generation sequencing (NGS), also called Massively Parallel Sequencing, to provide a confirmation and quantitation of MPS findings.
MPS in principle would be expected to overcome some of the limitations imposed by dPCR.
MPS provides comprehensive blood group profiling, relevant for blood donors and patients, in a single test system (35-37). In the case of NIPT, and provided sufficient sequencing depth is obtained, it would also permit inclusion of informative SNP targets to verify the presence of cffDNA in the sample. Whilst a review of MPS for NIPT application is beyond the scope of this review, some preliminary studies have been reported for a limited number of blood group markers (38,39). The key challenge with MPS will be, in contrast with dPCR, the management and validation of the bioinformatics data from a large number of target alleles.
Finally, NIPT studies to date have focussed on few antigens across multiple systems, particularly those more clinically relevant in the Caucasian population, such as the Rh blood group antigen, D (RH1) and the Kell blood group antigen, K (KEL1). The prevalence and significance of blood group antigens varies amongst population groups however. A recent case study highlights the potential for antigens prevalent in other populations to be implicated in HDFN. This particular case involved a pregnant woman, alloimmunized to GP Mur, where the fetus had a paternally-inherited GYP*Mur allele, GP. Mur, (GYP*501), within the MNS blood group system, is prevalent in Asian populations (40,41). The future opens the possibility for NIPT to provide a personalised precision medicine approach for such cases, using dPCR and potentially MPS platforms, to assist in early fetal blood group genotyping to guide pregnancy management.
Conclusions
There is a clinical need to measure the fetal blood group status from mothers who present with clinically significant red cell antibodies to guide antenatal care pathways (42).
The power of dPCR lies in the partitioning of DNA molecules, thereby removing competing high abundant from low abundant molecules.
Clinical feasibility studies to date, though small in number, are demonstrating that dPCR for fetal blood group genotyping provides an added diagnostic tool for the management of women with red cell antibodies. For the future, the potential for combining a dual approach of Massively Parallel Sequencing with dPCR awaits development.
Acknowledgments
Australian governments fund the Australian Red Cross Lifeblood to provide blood, blood products and services to the Australian community.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Frederik Banch Clausen) for the series “Blood Group Genotyping” published in Annals of Blood. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://aob.amegroups.com/article/view/10.21037/aob-22-4/coif). The series “Blood Group Genotyping” was commissioned by the editorial office without any funding or sponsorship. RLF serves as an unpaid editorial board member of Annals of Blood from April 2017 to April 2024. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lo YMD. Noninvasive prenatal testing: Advancing through a virtuous circle of science, technology and clinical applications. Prenat Diagn 2021;41:1190-2. [Crossref] [PubMed]
- Lo YM, Tein MS, Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet 1998;62:768-75. [Crossref] [PubMed]
- Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet 1997;350:485-7. [Crossref] [PubMed]
- Chan KC, Zhang J, Hui AB, et al. Size distributions of maternal and fetal DNA in maternal plasma. Clin Chem 2004;50:88-92. [Crossref] [PubMed]
- Finning K, Martin P, Daniels G. A clinical service in the UK to predict fetal Rh (Rhesus) D blood group using free fetal DNA in maternal plasma. Ann N Y Acad Sci 2004;1022:119-23. [Crossref] [PubMed]
- Colin Y, Chérif-Zahar B, Le Van Kim C, et al. Genetic basis of the RhD-positive and RhD-negative blood group polymorphism as determined by Southern analysis. Blood 1991;78:2747-52.
- Hyland CA, Gardener GJ, Davies H, et al. Evaluation of non-invasive prenatal RHD genotyping of the fetus. Med J Aust 2009;191:21-5. [Crossref] [PubMed]
- van der Schoot CE, Hahn S, Chitty LS. Non-invasive prenatal diagnosis and determination of fetal Rh status. Semin Fetal Neonatal Med 2008;13:63-8. [Crossref] [PubMed]
- de Haas M, Thurik FF, Koelewijn JM, et al. Haemolytic disease of the fetus and newborn. Vox Sang 2015;109:99-113. [Crossref] [PubMed]
- Sillence KA, Roberts LA, Hollands HJ, et al. Fetal Sex and RHD Genotyping with Digital PCR Demonstrates Greater Sensitivity than Real-time PCR. Clin Chem 2015;61:1399-407. [Crossref] [PubMed]
- O'Brien H, Hyland C, Schoeman E, et al. Non-invasive prenatal testing (NIPT) for fetal Kell, Duffy and Rh blood group antigen prediction in alloimmunised pregnant women: power of droplet digital PCR. Br J Haematol 2020;189:e90-4. [Crossref] [PubMed]
- Hyland CA, Millard GM, O'Brien H, et al. Non-invasive fetal RHD genotyping for RhD negative women stratified into RHD gene deletion or variant groups: comparative accuracy using two blood collection tube types. Pathology 2017;49:757-64. [Crossref] [PubMed]
- Tsui NB, Hyland CA, Gardener GJ, et al. Noninvasive fetal RHD genotyping by microfluidics digital PCR using maternal plasma from two alloimmunized women with the variant RHD(IVS3+1G>A) allele. Prenat Diagn 2013;33:1214-6. [Crossref] [PubMed]
- Nogués N. Recent advances in non-invasive fetal HPA-1a typing. Transfus Apher Sci 2020;59:102708. [Crossref] [PubMed]
- Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604-10. [Crossref] [PubMed]
- Quan PL, Sauzade M, Brouzes E. dPCR: A Technology Review. Sensors (Basel) 2018;18:1271. [Crossref] [PubMed]
- Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A 1999;96:9236-41. [Crossref] [PubMed]
- Fan HC, Quake SR. Detection of aneuploidy with digital polymerase chain reaction. Anal Chem 2007;79:7576-9. [Crossref] [PubMed]
- Lun FM, Chiu RW, Chan KC, et al. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin Chem 2008;54:1664-72. [Crossref] [PubMed]
- Wagner FF, Frohmajer A, Flegel WA. RHD positive haplotypes in D negative Europeans. BMC Genet 2001;2:10. [Crossref] [PubMed]
- Avent ND, Reid ME. The Rh blood group system: a review. Blood 2000;95:375-87.
- Gardener GJ, Legler TJ, Hyett JA, et al. Anti-D in pregnant women with the RHD(IVS3+1G>A)-associated DEL phenotype. Transfusion 2012;52:2016-9. [Crossref] [PubMed]
- Wagner T, Körmöczi GF, Buchta C, et al. Anti-D immunization by DEL red blood cells. Transfusion 2005;45:520-6. [Crossref] [PubMed]
- Körmöczi GF, Gassner C, Shao CP, et al. A comprehensive analysis of DEL types: partial DEL individuals are prone to anti-D alloimmunization. Transfusion 2005;45:1561-7. [Crossref] [PubMed]
- Svobodová I, Pazourková E, Hořínek A, et al. Performance of Droplet Digital PCR in Non-Invasive Fetal RHD Genotyping - Comparison with a Routine Real-Time PCR Based Approach. PLoS One 2015;10:e0142572. [Crossref] [PubMed]
- Gassner C, Castilho L, Chen Q, et al. International Society of Blood Transfusion Working Party on Red Cell Immunogenetics and Blood Group Terminology Report of Basel and three virtual business meetings: Update on blood group systems. Vox Sang 2022;117:1332-44. [Crossref] [PubMed]
- Storry JR, Clausen FB, Castilho L, et al. International Society of Blood Transfusion Working Party on Red Cell Immunogenetics and Blood Group Terminology: Report of the Dubai, Copenhagen and Toronto meetings. Vox Sang 2019;114:95-102. [Crossref] [PubMed]
- Hyland CA, O'Brien H, Flower RL, et al. Non-invasive prenatal testing for management of haemolytic disease of the fetus and newborn induced by maternal alloimmunisation. Transfus Apher Sci 2020;59:102947. [Crossref] [PubMed]
- Li S, Mo C, Huang L, et al. Hemolytic disease of the fetus and newborn due to alloanti-M: three Chinese case reports and a review of the literature. Transfusion 2019;59:385-95. [Crossref] [PubMed]
- Pal M, Williams B. Prevalence of maternal red cell alloimmunisation: a population study from Queensland, Australia. Pathology 2015;47:151-5. [Crossref] [PubMed]
- D'Aversa E, Breveglieri G, Pellegatti P, et al. Non-invasive fetal sex diagnosis in plasma of early weeks pregnants using droplet digital PCR. Mol Med 2018;24:14. [Crossref] [PubMed]
- Poole J, Daniels G. Blood group antibodies and their significance in transfusion medicine. Transfus Med Rev 2007;21:58-71. [Crossref] [PubMed]
- Markham KB, Rossi KQ, Nagaraja HN, et al. Hemolytic disease of the fetus and newborn due to multiple maternal antibodies. Am J Obstet Gynecol 2015;213:68.e1-5. [Crossref] [PubMed]
- Hendrickson JE, Delaney M. Hemolytic Disease of the Fetus and Newborn: Modern Practice and Future Investigations. Transfus Med Rev 2016;30:159-64. [Crossref] [PubMed]
- Schoeman EM, Lopez GH, McGowan EC, et al. Evaluation of targeted exome sequencing for 28 protein-based blood group systems, including the homologous gene systems, for blood group genotyping. Transfusion 2017;57:1078-88. [Crossref] [PubMed]
- Schoeman EM, Roulis EV, Liew YW, et al. Targeted exome sequencing defines novel and rare variants in complex blood group serology cases for a red blood cell reference laboratory setting. Transfusion 2018;58:284-93. [Crossref] [PubMed]
- Roulis E, Schoeman E, Hobbs M, et al. Targeted exome sequencing designed for blood group, platelet, and neutrophil antigen investigations: Proof-of-principle study for a customized single-test system. Transfusion 2020;60:2108-20. [Crossref] [PubMed]
- Rieneck K, Bak M, Jønson L, et al. Next-generation sequencing: proof of concept for antenatal prediction of the fetal Kell blood group phenotype from cell-free fetal DNA in maternal plasma. Transfusion 2013;53:2892-8. [Crossref] [PubMed]
- Wienzek-Lischka S, Krautwurst A, Fröhner V, et al. Noninvasive fetal genotyping of human platelet antigen-1a using targeted massively parallel sequencing. Transfusion 2015;55:1538-44. [Crossref] [PubMed]
- Mallari RA, Chan A, Powers RJ, et al. Fetal inheritance of GP*Mur causing severe HDFN in an unrecognized case of maternal alloimmunization. Transfusion 2020;60:870-4. [Crossref] [PubMed]
- Lopez GH, Hyland CA, Flower RL. Glycophorins and the MNS blood group system: a review. Ann Blood 2021;6:39.
- Dziegiel MH, Krog GR, Hansen AT, et al. Laboratory Monitoring of Mother, Fetus, and Newborn in Hemolytic Disease of Fetus and Newborn. Transfus Med Hemother 2021;48:306-15. [Crossref] [PubMed]
Cite this article as: Hyland CA, O’Brien H, McGowan EC, Perros AJ, Flower RL, Lopez GH, Gardener GJ. The power of digital PCR in fetal blood group genotyping: a review. Ann Blood 2023;8:6.


