
RH genetic variation and the impact for typing and personalized transfusion strategies: a narrative review
Introduction
Rationale
The purpose of this narrative review is to provide the reader with updated guidance on the clinical utility identifying patients at risk for allo-anti-D formation as well as strategies that can be used to determine candidacy for Rh immune globulin in pregnant women. This narrative also reviews the genetic complexity behind phenotypes lacking high prevalence antigens encoded by the RHCE gene and describes an effective strategy that has been used for more than a decade in the US to personalize donor unit selection in Rh alloimmunized patients based on RH allele information. This strategy can be used in alloimmunized patients in whom transfusion has become seemingly impossible. The objective is to provide the reader with information that can be used to reduce Rh alloimmunization and increase personalized patient care related to red cell alloimmunization. I present the following article in accordance with the Narrative Review reporting checklist (available at https://aob.amegroups.com/article/view/10.21037/aob-22-6/rc).
Methods
Peer-reviewed literatures published in English and listed in PubMed from 1960 to January 2022 were reviewed while compiling this work. In addition, AABB guidance documents and AABB standards as well as guidelines published by the American Society of Hematology and the British Society of Haematology were reviewed and cited where appropriate. The allele tables on the website of the ISBT Working Party for Red Cell Immunogenetics and Blood Group Terminology were also reviewed and referenced. Relevant peer-reviewed abstracts by the author were reviewed and compiled data from such work is included where appropriate (see Table 1).
Table 1
| Items | Specification |
|---|---|
| Date range of searches | November 2021 to January 2022 |
| Databases and other sources searched | PubMed; peer-reviewed abstracts published or co-published by the author; service requests received from the American Red Cross National Molecular Laboratory from July 2018 to June 2021; the ISBT Working Party on Red Cell Immunogenetics and Blood Group Terminology RHD and RHCE allele tables |
| Terms used to search PubMed | “RhD”[MeSH] AND “discrepan*”[MeSH] |
| “RhD”[MeSH] AND “alloimmun*”[MeSH] | |
| “Rh”[MeSH] AND “matching”[MeSH] | |
| “RhD”[MeSH] AND “weak”[MeSH] | |
| “RhD”[MeSH] AND “partial”[MeSH] | |
| “sickle”[MeSH] AND “prophylactic”[MeSH] AND “antigen”[MeSH} | |
| “American Society of Hematology”[MeSH] AND “sickle”[MeSH] AND “guidance”[MeSH] | |
| “British Society of Hematology”[MeSH] AND “sickle”[MeSH] AND “guidance”[MeSH] | |
| “American Rare Donor Program”[MeSH] | |
| Timeframe | 1960–January 2022 |
| Inclusion and exclusion criteria | Focus was placed on original papers and reviews in English about the RH blood group system, with specific focus on Rh variants, serologic discrepancies and population studies. |
| Selection process | It was conducted independently by the author |
The RH blood group system
The RH blood group system includes two genes (RHD and RHCE), both with 10 exons, that encode a total of 56 antigens. While the RHD gene encodes one common antigen, RhD or RH1, the RHCE gene encodes two sets of antithetical common antigens, big C (RH2) and little c (RH4) and big E (RH3) and little e (RH5). Expression of RhD and RhCE antigens requires co-expression with the RhAG protein in a multimeric protein complex.
RH locus
The RH locus, located on Chromosome 1, includes the highly homologous and highly polymorphic RHD and RHCE genes. The RhD and Rhce polypeptides are multipass transmembrane proteins that are part of a multiprotein complex on the red cell surface. The RhD polypeptide carries the RhD antigen with multiple epitopes and can express neoepitopes such as Goa (RH30) or DAK (RH54). The RHCE gene has four normal allele types (RHCE*ce, RHCE*Ce, RHCE*cE and RHCE*CE) that encode Rhce polypeptides expressing antithetical antigens c or C and e or E.
The RHD gene
The RHD gene (NG_007494) includes 10 exons and encompasses ~58,000 base pairs of DNA on chromosome 1. It is polymorphic with variants including deletions, gene conversions, and non-synonymous single nucleotide variants (1).
There are several mechanisms and more than 80 RHD alleles that are associated with a null or RhD negative phenotype (see Table 2). RHD*01N.01, deletion of the gene’s 10 exons is the most common RhD negative allele in Caucasians while RHD*08N.01, the silenced RHD allele is the most common RhD negative allele in individuals of African descent. There are several RHD alleles (RHD*01N.02 – RHD*01N.05) that do not express the D antigen due to gene conversion involving replacement of multiple exons with those from the RHCE gene. In addition, several RHD alleles involve gene conversion that abolishes RhD expression and gains expression of a variant of the C antigen, with RHD*DIIIa-CE(4-7)-D or RHD*03N.01 the most common in African Americans. Finally, there are RHD alleles in which splicing defects abolish RhD expression (e.g., RHD*01N.24), those with insertion/deletion polymorphisms that lead to frameshifts and premature termination (ex. RHD*01N.11) and those with nonsense mutations (ex. RHD*01N.18) as well as those with nonsynonymous mutations where the mechanism is less clear (ex. RHD*01N.80).
Table 2
| Race/ethnicity | n (%) |
|---|---|
| AA | 363 (41.3) |
| Cauc | 366 (41.6) |
| Other | 98 (11.1) |
| Hisp | 28 (3.2) |
| NP | 10 (1.1) |
| MR | 6 (0.7) |
| API | 5 (0.6) |
| AIAN | 3 (0.3) |
AA, African American; Cauc, Caucasian; Hisp, Hispanic; NP, not provided; MR, mixed race; API, Asian Pacific Islander; AIAN, American Indian Alaska Native.
RhD antigen
The RhD polypeptide expresses the RhD or RH1 antigen. RHD alleles that encode RhD antigens have historically been categorized on one of three groups (weak, partial or DEL). Partial RhD antigens are defined as antigens in which one or more epitopes recognized by monoclonal antibodies is lacking.
As is the case with some other blood group antigens, including other antigens in the RH blood group system, expression is not binary, meaning that classifying the phenotype as positive or negative is incomplete and inaccurate. Though the red cells may test positive with an anti-D reagent, if the RHD gene sequence differs from the consensus sequence the antigen may be different. These differences may impact copy number, gain or loss of antigen epitopes or both as well as reactivity with human or reagent anti-D. Panels of monoclonal antibodies have been used to investigate the epitope loss in RhD variants (2) and flow cytometry has been used to assess D antigen sites per red blood cell (RBC) in subjects expressing RhD variants (3).
Changing terminology
In 1946, the weak D phenotype was initially described and defined as red cells that agglutinate with some but not all anti-D reagents and was given the term DU (4). When anti-D sensitizes the RBCs but they do not directly agglutinate, visualization can be enhanced by the use of antiglobulin sera. Subsequently, the US put in place policies requiring blood donor RBCs that type D- to be retested using antiglobulin (“weak D test”) and if agglutination was seen, to be classified as D+ (5). This additional testing is not required of pregnant women who type D-; instead, they are given Rh immunoglobulin (RhIg). The term “D mosaic” was coined in 1962 to describe red cells that express RhD antigen that is missing one or more epitopes defined by reactivity with monoclonal anti-D antisera (6). While initially the DU term was used to define a red cell with a quantitative but not qualitative difference in the D antigen, this terminology was fraught with challenges. First because there is no DU antigen or anti-DU antibody. Second because some DU+ individuals, initially thought to carry little risk of allo-anti-D formation, were found to make allo-anti-D. The term DU was replaced with “weak D” in 1992 (7).
Changing guidance
Since the 1990s, our understanding of the complexity of RHD genetic variation and its impact on the RhD antigen has grown significantly. In 2013, Geoffrey Daniels proposed using the term “D variant” to avoid confusion with antigens that can type weakly D+, yet are associated with epitope loss and allo-anti-D production (8). In 2014, Sandler et al. presented results of a College of American Pathology (CAP) survey of 3,100 US hospitals about policies and procedures around testing individuals with weak D phenotypes and administration of RhIg which highlighted the lack of standard practice in the US (9). In 2015, an inter-organizational work group published a commentary (10) with two key take aways, both illustrated in Figure 1.
- A serologic weak D phenotype was redefined as reactivity of RBCs with an anti-D reagent giving no or weak (≤2+) reactivity in initial testing, but agglutinating moderately or strongly with antihuman globulin.
- The RhD phenotype of individuals with a serologic weak D phenotype should be resolved by using RHD genotyping to identify the RHD alleles carried by the patient. In this way, individuals found to carry RHD variant alleles weak D type 1, 2 or 3 (RHD*01W.1, RHD*01W.2, RHD*01W.3) are not considered at risk for allo-anti-D (10), and would avoid RhIg administration.
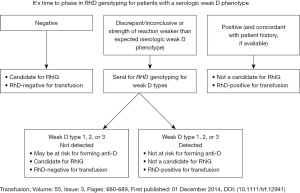
Based on the number of live births and transfusion recipients and ethnic background of the US population, this strategy was estimated to identify more than 13,000 women of child-bearing age in whom more than 24,000 doses of RhIg would not be needed. The recommendation to “phase-in” RHD genotyping, specifically of women of child-bearing age with a serologic phenotype was endorsed by the AABB and CAP (11). Financial modeling suggested that this approach could add clinical benefit without additional costs (12).
Strategies for identifying those at risk
Since the release of the joint statement (11), there has been a growing appreciation for the ability of molecular methods to assess a patient’s risk of alloimmunization to RhD more precisely. However, besides the flow chart from Sandler et al. (10) reproduced in Figure 1 and a review (13), there is no specific guidance for how a hospital blood bank would identify patients that may benefit from RHD genotyping. Further, many hospital blood banks have automated antigen typing platforms, whether solid phase or gel technology and these platforms may or may not give the user the ability to easily review results of specimens demonstrating weaker reactivities. Several healthcare systems have evaluated algorithms to select samples for RHD genotyping. A repeating theme is the use of differential anti-D reactivity between two methods or reagents to identify those who likely carry RHD variant alleles and express RhD variant antigens. It is important to emphasize that serologic testing cannot accurately distinguish between RhD variant antigens that put the patient at risk of alloimmunization or not. RHD genotyping of these individuals can identify the specific alleles and that information can be used to assess risk of alloimmunization. This strategy can focus genotyping efforts on patients who would benefit the most, since it is not currently cost-effective to perform RHD genotyping of all patients and blood donors.
There have been several publications that describe the use of anti-D reactivity differences to identify patients who could benefit from RHD genotyping (14-19) (see Table 3). These studies share a two reagent and/or two-method strategy and are highly effective at identifying samples for which RHD genotyping predicts expression of RhD variants. Importantly, monoclonal anti-D panels are now recognized as being ineffective at differentiating patients at risk of allo-anti-D while RHD genotyping can differentiate samples carrying weak D types 1, 2 and 3 from samples carrying partial D alleles. The two-test approach does not require additional testing in laboratories accredited by AABB, since these organizations already require two determinations of ABO/Rh for pre-transfusion testing for allogeneic transfusion (31).
Table 3
| First author (reference) | Subject No. |
Patient or donor |
RHD variant alleles | % of total |
Weak Type 1, 2, or 3 | % of variants |
Weak Type 4.0 |
% of variants |
Distinct alleles found |
|---|---|---|---|---|---|---|---|---|---|
| USA | |||||||||
| Keller (20) | 879 | P | 558 | 63 | 277 | 50 | 87 | 16 | 19 |
| Wang (21) | 11 | P | 10 | 91 | 1 | 10 | 4 | 40 | 4 |
| Hudgins (19) | 80 | P | 52 | 65 | 10 | 19 | 20 | 38 | 15 |
| Canada | |||||||||
| Denomme (14) | 55 | P | 53 | 96 | 25 | 47 | 6 | 11 | 12 |
| Brazil | |||||||||
| Campos (22) | 129 | P | 98 | 76 | 83 | 85 | 14 | 14 | 6 |
| Bub (23) | 104 | P | 104 | 100 | 23 | 22 | 51 | 49 | 9 |
| Dezan (24) | 58 | P | 58 | 100 | 29 | 50 | 0 | 0 | 4 |
| Iran | |||||||||
| Oodi (25) | 100 | P&D | 100 | 100 | 15 | 15 | 0 | 0 | 18 |
| Thailand | |||||||||
| Thongbut (26) | 51 | D | 38 | 75 | 0 | 0 | 0 | 0 | 13 |
| Korea | |||||||||
| Chung (27) | 23 | P | 21 | 91 | 0 | 0 | 0 | 0 | 10 |
| Egypt | |||||||||
| Bakry (28) | 50 | P&D | 45 | 90 | 15 | 33 | 11 | 24 | 10 |
| China | |||||||||
| Yan (29) | 32 | D | 32 | 100 | 0 | 0 | 0 | 0 | 9 |
| Tunisia | |||||||||
| Ouchari (30) | 67 | D | 64 | 96 | 2 | 3 | 60 | 94 | 5 |
Benefit of RHD genotyping: avoiding overtreatment
Among Caucasians, in whom a serologic weak D phenotype is found at a prevalence of 0.2–1% (15,32), 73–80% of these will be found to carry RHD alleles weak D type 1, 2 or 3 (RHD*01W.1, RHD*01W.2, RHD*01W.3) (10,15). Thus, in a primarily Caucasian population, the main benefit of implementing a process to identify patients with serologic weak D phenotype for RHD genotyping will be identification of those women of childbearing age who do not require RhIg prophylaxis. Importantly, the benefits and the outcome of such an approach may be different in other ethnic groups.
Benefit of RHD genotyping: identifying those at risk who are hiding in plain sight
Among non-Caucasians, in whom a serologic weak D phenotype is more often associated with inheritance of a partial D allele, implementing an RHD genotyping strategy can identify patients who may have typed D+ on an automated testing instrument and who may not have been identified as at risk of allo-anti-D otherwise. Though some monoclonal anti-D reagents will be non-reactive with red cells expressing a partial D phenotype, it is common for such cells to react strongly with some anti-D reagents typically used on automated platforms in the US yet weakly or not at all by manual tube agglutination method. Implementing a process to test women of child-bearing age by two different methods and/or anti-D reagents will help to identify women who may express a partial D phenotype and who could benefit from RHD genotyping to accurately assess candidacy for RhIg. A prerequisite for this approach is prenatal care with access to laboratory testing in a timeframe that allows for appropriate RhIg administration during and shortly after delivery.
Findings of a large national US reference laboratory
Keller et al. (20) described RHD genotyping of 879 patientsamples referred to a large national molecular laboratory from across the US in calendar year 2020. These specimens were identified by hospitals based on their own criteria, which were not provided. The race/ethnicity of the cohort was 41% African American (AA), 41% Caucasian (CAUC), 11.1% “other”, 3.2% Hispanic, 1.1% race not provided, 0.7% mixed race, 0.6% Asian and 0.3% Native American. RHD genotyping predicted 63.5% to express a D variant with 31.5% hemizygous or homozygous for Weak D Types 1, 2 and 3. Among Caucasian patients, the majority (61.7%) carried Weak D Types 1, 2 and 3. Among African American patients, 58% were predicted to express a partial D phenotype with 16 different alleles identified, of which the most common RHD variant allele was weak D type 4.0 (Table 2). The RHD alleles identified in this large US cohort are listed in Table 4. The breakdown of patients by race/ethnicity that were found to carry Weak D Types 1, 2 or 3 or other RHD alleles encoding partial D are listed in Table 5 and Table 6. This study is the largest of several published studies (Table 3). Approximately two-thirds (63%) of subjects were found to carry a variant RhD antigen. This percentage is lower than the other studies where subjects were identified using a specific algorithm, such as that used in Luo et al. or Horn et al. (17,18). Interestingly, when the large study (20) is compared to Hudgins et al. (19), in which one-tenth the number of subjects were tested, a similar percent of RHD variants were identified (65% vs. 63%) and though the sample size of Keller et al. was more than ten-fold larger than that of Hudgins et al., it identified 19 distinct RHD variant alleles while Hudgins identified 15 alleles. This may suggest that the number of alleles involved in typing discrepancies in the US population is small and mostly known.
Table 4
| RHD allele# | All patients (n=879) | Caucasians (n=366) | African Americans (n=363) | Other (n=150) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | ||||
| RHD*01 | 245 | 27.9 | 94 | 25.7 | 104 | 29 | 47 | 31.3 | |||
| RHD*01W.01 (weak D Type 1) | 149 | 17.0 | 123 | 33.6 | 4 | 1.1 | 22 | 14.7 | |||
| RHD*01W.02 (weak D Type 2) | 100 | 11.4 | 77 | 21.0 | 6 | 1.7 | 17 | 11.3 | |||
| RHD*weak D Type 4.0 | 87 | 9.9 | 12 | 3.3 | 67 | 18.5 | 8 | 5.3 | |||
| RHD*01W.03 (weak D Type 3) | 30 | 3.4 | 28 | 7.7 | 1 | 0.3 | 1 | 0.7 | |||
| RHD*DAR | 71 | 8.1 | 2 | 0.5 | 54 | 14.9 | 15 | 10 | |||
| RHD*DIIIa | 17 | 1.9 | 0 | 0 | 17 | 4.7 | 0 | 0 | |||
| RHD*DIVa | 12 | 1.4 | 0 | 0 | 11 | 3.0 | 1 | 0.7 | |||
| RHD*08N.01 (RHD*Psi) | 5 | 0.6 | 0 | 0 | 5 | 1.4 | 0 | 0 | |||
| RHD*DOL | 4 | 0.5 | 1 | 0.3 | 2 | 0.6 | 1 | 0.7 | |||
| RHD*DAU5 | 4 | 0.5 | 0 | 0 | 3 | 0.8 | 1 | 0.7 | |||
| RHD*DCS or DFV | 3 | 0.3 | 2 | 0.5 | 0 | 0 | 1 | 0.7 | |||
| RHD*DFR | 3 | 0.3 | 1 | 0.3 | 0 | 0 | 2 | 1.3 | |||
| RHD*DAU0 | 2 | 0.2 | 0 | 0 | 2 | 0.6 | 0 | 0 | |||
| RHD*DWN | 2 | 0.2 | 0 | 0 | 2 | 0.6 | 0 | 0 | |||
| RHD*DAU4 | 1 | 0.1 | 0 | 0 | 1 | 0.3 | 0 | 0 | |||
| RHD*DUC | 1 | 0.1 | 1 | 0.3 | 0 | 0 | 0 | 0 | |||
| RHD*DIIIc | 1 | 0.1 | 0 | 0 | 0 | 0 | 1 | 0.7 | |||
| RHD*DVI | 1 | 0.1 | 0 | 0 | 0 | 0 | 1 | 0.7 | |||
| RHD*DNB | 1 | 0.1 | 1 | 0.3 | 0 | 0 | 0 | 0 | |||
| RHD*DSPM | 1 | 0.1 | 0 | 0 | 1 | 0.3 | 0 | 0 | |||
| RHD*667G,809G | 1 | 0.1 | 1 | 0.3 | 0 | 0 | 0 | 0 | |||
#, Homozygosity and hemizygosity due to RHD*01N.01 were not differentiated. RHD*03N.01 alleles cannot be quantified since allele assignments in many cases were equivocal [ex. DIIIa/DIIIa or DIIIa/DIIIa-CE(4-7)-D].
Table 5
| RHD allele | Risk of allo-anti-D |
All patients | Caucasian | African American | Other | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | |||||
| RHD*01 | N | 281 | 32 | 97 | 26.5 | 130 | 35.8 | 27 | 27.6 | |||
| Weak D Type 1, 2 or 3 | N | 277 | 31.5 | 226 | 61.7 | 11 | 3 | 30 | 30.6 | |||
| Weak D Type 4 | ?? | 87 | 9.9 | 12 | 3.3 | 67 | 18.5 | 4 | 4.1 | |||
| Various partial D alleles | Y | 194 | 22.1 | 11 | 3 | 141 | 39.7 | 31 | 31.6 | |||
| RHD*01N.01 | Y | 40 | 4.6 | 20 | 5.5 | 11 | 3 | 6 | 6.1 | |||
N, no; Y, yes; ??, no consensus.
Table 6
| RHD alleles | All patients | Caucasian | AA (n) | API (n) | Hisp (n) | AIAN (n) | Other (n) | Mixed (n) | NP (n) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | |||||||||
| Weak D Type 1, 2 or 3 | 277 | 31.5 | 226 | 61.7 | 11 | 1 | 3 | 2 | 30 | 0 | 1 | |
| Type 1 | 147 | 16.7 | 121 | 33.1 | 4 | 1 | 3 | 0 | 17 | 0 | 1 | |
| Type 2 | 98 | 11.1 | 75 | 20.5 | 6 | 0 | 0 | 2 | 12 | 0 | 0 | |
| Type 3 | 30 | 3.4 | 28 | 7.7 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | |
| Type 1/Type 2 | 2 | 0.2 | 0 | N/A | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
AA, African American; API, Asian Pacific Islander; Hisp, Hispanic; AIAN, American Indian Alaska Native; NP, not provided; N/A, not applicable.
Phasing out serologic weak D result
In 2020, Flegel et al. published a commentary (33) that suggested that blood banks and Immunohematology reference laboratories should stop releasing reports with “weak D+” or “serologic weak D+” interpretations as a final result. Instead, the authors suggest that such cases should be referred to molecular reference laboratories where genotyping can be used to identify the RHD alleles. With knowledge of the RHD alleles that are responsible for the weak D phenotype, the risk of alloimmunization can be more accurately assessed. In pregnant women and women of child-bearing age, this information is critical to determine candidacy for RhIG; in all patients, the information can be used to determine which require RhD negative blood products. This process offers a genome-informed and personalized approach to RhD alloimmunization risk assessment. Furthermore, since RHD alleles associated with partial D phenotypes are often co-inherited with RHCE alleles encoding partial antigens or lacking high prevalence antigens, this information may uncover additional Rh alloimmunization risk. The American Red Cross National Molecular Laboratory saw a 6.3% increase in service requests for RHD genotyping of patients from FY19 (July 2018 to June 2019) to FY2020 (July 2019 to June 2020) followed by a 23.8% increase from FY2020 to FY2021 (July 2020 to June 2021). The later increase may have been influenced by the Flegel et al. publication (33) which was released electronically in March 2020.
Personalized transfusion support for patients lacking high prevalence antigens in the RH system
In patient populations where chronic transfusion is a therapeutic modality (ex. sickle cell anemia and thalassemia), RBC phenotype matching has been used for more than twenty-five years (34-37). More recently, prophylactic red cell antigen matching for common RBC antigens has become commonplace (38,39). Matching for C, E and K is recommended by the British Society of Haematology (40). The American Society of Hematology (ASH) recently recommended matching for C, E and K as the standard of care for all patients with sickle cell disease, optimally before the first transfusion (41). The ASH guidelines also recommend that when RH genotyping identifies a partial C antigen (either by RHD*03N.01 or RHCE*Ce.10) in a patient without a conventional RHCE allele expressing the C antigen, the patient should receive C- red cell products to avoid allo-anti-C. In addition, genotyping for red cell antigens is preferred over serologic phenotyping due to increased accuracy and the ability to predict phenotypes for antigens for which commercial reagents are not available or not reliable (41-43).
Personalized transfusion support for patients with sickle cell disease
In patients with SCD, it has been appreciated for some time that since this population is predominantly of African descent, and since in the USA the blood donor base is primarily Caucasian, there is a high likelihood of mismatches in what had been termed “minor” blood group antigens (i.e., Fya, Fyb, Jka, Jkb, S, s). In the US, the desire to “extend” the match from ABO/Rh and C and E antigens to these additional antigens resulted in a variety of donor recruitment programs aimed at collecting RBC units from African American donors (44). However, individuals of African descent have extensive genetic variation in the RH locus, with not only partial RhD phenotypes but loss of high prevalence antigens such as hrB, hrS and Hr (45). While RBC units from Caucasians with normal Rh antigens may put patients of African descent with partial Rh antigens at risk of alloantibodies in the RH system, RBC units from African descent donors expressing neoepitopes such as Goa in individuals with RHD*DIVa can alloimmunize patients of African descent who do not express the same RhD variant antigen(s) (46). A study of Brazilian patients with SCD found that those who typed D+ with unexpected anti-D carried RHD alleles encoding partial D antigens (47). Additionally, patients of African descent who type C+ with anti-C and/or e+ with anti-e often carry variant RHCE alleles encoding partial antigens. In the case of the C+ patient with anti-C, they may not express the C antigen but instead a hybrid RHD-RHCE allele that encodes a partial C antigen (48).
There has been increasing interest in molecular matching for patients with SCD, including matching at the allele level for RH variants (49). In 2017, Fasano et al. (50) noted that more studies would be needed to determine if use of RH-allele matched RBC units would be a feasible approach to prevent Rh alloimmunization. The feasibility of such an approach was later examined using virtual simulations (51). The study involved use of identical matches, haplotype matches and less restrictive matches that classified RHD*DAU0 as equivalent to RHD*01 and RHCE*ce48C equivalent to RHCE*ce. A recent modeling study provides evidence that some RhD variants may be “benign” based on impact of amino acid substitutions on tertiary structure of the RhD molecule (52). Chou et al. (51) noted that the success of prophylactic RH allele matching protocol would be dependent on improved recruitment of African American blood donors, an issue also highlighted by Karafin et al. (53).
Identifying patients lacking hrB (RH31) and/or hrS (RH19)
There are multiple RHCE alleles that predict loss of high-prevalence antigens hrB (RH31) and/or hrS (RH19) (42) (Table 7). This is an important distinction from most other blood group antigens and should be taken into account when designing a process for selection of RBC units based on RH alleles and their predicted phenotypes. In addition, there are no commercial reagents to screen for donors lacking either of these antigens such that molecular screening is the primary means of donor identification. While many of the RHCE*ce alleles that encode Rhce molecules lacking hrB include the single nucleotide variant (SNV) C>G at coding position 733 in the RHCE gene (c.733C>G), not all alleles carrying c.733G are associated with an hrB- phenotype and for those that do, most but not all will express both V and VS, since the V is lost when the c.733G is coinherited with c.1006T. And there are RHCE*ce alleles such as RHCE*ceAG that do not carry c.733G yet encode an hrB- phenotype. The RHCE*ce alleles that encode hrS- phenotype are varied in the SNVs they carry; several carry c.712 and some gain expression of the low prevalence antigen RH49 (STEM). This heterogeneity requires, at minimum, medium-resolution RHCE genotype information for selection of donor units based on RH alleles of the donor and patient. While the commonly used commercial red cell genotyping kits are low-resolution, containing a small number of markers that are insufficient to assign alleles with confidence, additional testing and/or medium-resolution assays include additional SNVs that allow allele assignments with increased accuracy (54). High-resolution methodologies such as Sanger sequencing of genomic DNA or cDNA or Next Generation sequencing provide the greatest accuracy, but analysis and interpretation are labor-intensive and complex (55).
Table 7
| RHCE allele (ISBT) | RHCE allele (alias) | RH31 (hrB) | RH19 (hrS) | RH18 (Hr) | RH10 (V) | RH20 (VS) | RH5 (e) | RH4 (c) | RH43 (Crawford) | RH49 (STEM) | RH58 (CELO) | RH59 (CEAG) | RH61 (CEVF) | RHD alleles often linked |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RHCE*ce.04.01 | RHCE*ceAR | pos | neg | neg | weak | neg | Partial | Partial | neg | neg | pos | pos | pos | RHD*DAR |
| RHCE*ce.05.01 | RHCE*ceEK | pos | neg | neg | neg | neg | Partial | Partial | neg | neg | pos | pos | pos | RHD*01N.01 |
| RHCE*ce.06.01 | RHCE*ceAG | neg | pos | pos | neg | neg | Partial | pos | neg | neg | pos | neg | pos | RHD*01N.01 |
| RHCE*ce.07.01 | RHCE*ceMO.01 | neg | neg | pos | neg | neg | Partial | Partial | neg | neg | pos | pos | neg | RHD*DAU |
| RHCE*ce.07.02 | RHCE*ceMO.02 | neg | neg | pos | neg | neg | Partial | Partial | neg | neg | pos | pos | neg | RHD*DAU |
| RHCE*ce.08 | RHCE*ceBI | pos | neg | neg | neg | neg | Partial | pos | neg | pos | pos | pos | pos | RHD*DOL |
| RHCE*ce.09 | RHCE*ceSM | pos | neg | neg | neg | neg | Weak | pos | neg | pos | pos | pos | pos | RHD*DOL |
| RHCE*ce.20.01 | RHCE*ce733G | vw/neg | pos | pos | pos | pos | Partial | Partial | neg | neg | pos | pos | pos | RHD*01 |
| RHCE*ce.20.02.01 | RHCE*48C,733G | neg | pos | pos | pos | pos | Partial | Partial | neg | neg | pos | pos | pos | RHD*weak D type 4.0 |
| RHCE*ce.20.03 | RHCE*ce48C,733G,1006T | neg | pos | pos | neg | pos | Partial | Partial | neg | neg | pos | pos | pos | RHD*DIIIa or RHD*DIIIa-CE(4-7)-D |
| RHCE*ce.20.04 | RHCE*ce48C,733G,1025T | neg | pos | pos | pos | pos | Partial | pos | neg | neg | pos | pos | pos | RHD*weak D type 4.0 or RHD*DIIIa |
| RHCE*ce.20.05 | RHCE*ce733G,1006T | neg | pos | pos | neg | pos | Partial | pos | neg | neg | pos | pos | pos | RHD*DIIIa-CE(4-7)-D |
| RHCE*ce.20.06 | RHCE*ceCF | neg | neg | pos | unk | pos | Partial | Partial | pos | neg | neg | pos | pos | RHD*weak D type 4.0 or RHD*01N.01 |
| RHCE*ce.20.08 | RHCE*48C,733G,748A | neg | pos | pos | pos | pos | Weak | pos | neg | neg | pos | pos | pos | RHD*weak D type 4.0 |
| RHCE*ce.33 | RHCE*506C | neg | pos | pos | neg | neg | Partial | pos | neg | neg | pos | pos | pos | RHD*weak D type 4.0 |
ISBT, International Society of Blood Transfusion.
Personalized transfusion support for alloimmunized patients using RH allele selection
The American Rare Donor Program (ARDP) aids in identifying rare blood products for alloimmunized patients (56). These products include units lacking high prevalence antigens hrB and hrS (defined by genotype) in the RH blood group system. In a review of ARDP activities from 2005 and 2010 (56), there were 10 requests for hrB- or hrS-units in 2005, and since 2010 these requests are filled with RH genotype-selected units, when possible (56,57). In 2013, the process or RH allele selection used by the ARDP was formalized (58) and additional reports of its use have been presented (59). In 2020, the ARDP received 126 requests for RH allele selected red cell units for 53 patients. An analysis of these requests (60) showed that 49% of these patients carried at least one RHCE*ce733G allele. Though the ISBT Working Party on Red Cell Immunogenetics and Blood Group Terminology currently classifies this allele as having an hrB+vw/- phenotype, allo-anti-hrB has been reported (61,62). Therefore, ARDP considers this allele to be associated with risk of allo-anti-hrB and provides RH allele selected units upon request.
As Chou et al. noted (51), large numbers of well characterized AA donors would be needed to put into practice routine selection of donor units based on RH alleles. To that end, the American Red Cross National Molecular Laboratory developed high-throughput assays to test E-, K-, hemoglobin (Hgb) S negative donors of African descent for both RHCE (63) and RHD (64) variants. In an analysis of more than 6,000 predominantly African American donors tested by HemoID DQS red cell genotyping panel (Agena Bioscience), 564 donors predicted to be E-, K-, HgbS- and predicted to be hrB+vw/- or hrB- or hrS negative or E-e+w were selected for further characterization. Genotyping was performed in 96-well format with custom RHCE (10 SNVs) and RHD (18 SNVs) oligonucleotide extension assays designed for MALDI-TOF. Table 8 shows that 82 (14.5%) of those characterized were predicted to be homozygous for RHCE*ce733G and predicted to be hrB+vw/-. All of these carried a normal RHD*01 allele. While 5% were predicted to be hrB- and only 0.5% predicted to be hrS-. It has not been well appreciated that the E- hrS- phenotype is at least an order of magnitude rarer than the E- hrB- phenotype. Also of note, nearly half of the hrB- and hrs- donors were predicted to express a partial D phenotype. There is a strong association between loss of the high prevalence antigens hrB and/or hrS and a partial D phenotype. In addition, individuals who are found to carry the RHCE*ce48C,733G,1006T allele may carry the RHD*DIIIa-CE(4-7)-D allele and together are associated with the r’s phenotype. This phenotype is associated with expression of a partial C antigen and in patients who do not express a normal C antigen, with risk of allo-anti-C. Another important factor derived from RHD and RHCE co-inheritance data is the finding that most RHCE*ce733G alleles are linked to a normal RHD*01 allele encoding a D+ phenotype.
Table 8
| Test or phenotype | N | Of all donors tested | Of those selected for RH characterization |
|---|---|---|---|
| RBC genotyping panel | 6,392 | – | – |
| Custom RHCE and RHD assays | 564 | 8.8% | – |
| Predicted to type hrB+vw/- | 82 | 1.3% | 14.5% |
| Predicted to type hrB- | 28 | 0.4% | 5.0% |
| r’s homozygous | 7 | 0.1% | 1.2% |
| Predicted to be hrS- | 3 | 0.05% | 0.5% |
| Predicted to express altered C | 44 | – | 7.8% |
| Predicted to express partial D | 268 | – | 47.5% |
RBC, red blood cell.
The RH allele selection process
Importantly, precise RH allele selection requires genotype information for RHCE SNVs not interrogated by commonly used low-resolution, commercially-available red cell genotyping panels. The ARDP requires RHCE c.48G>C, c.340C>T, c.697C>G, c.712A>G, c.733C>G, c.1006G>T and c.1025C>T genotype results for submission of a rare donor based on hrB- phenotype and RHCE c.48G>C, c.667G>T, c.712A>G, 818C>T and 916A>G for submission of a rare donor based on hrS- phenotype. Since most laboratories are not performing high-resolution testing, the alleles assigned to the patient are considered the “probable alleles” to convey that there is the potential that the patient carries other variants not interrogated that may change the allele assignment and, in some cases, the predicted phenotype. It is also noteworthy that with more laboratories performing high-resolution methods like full gene or exon sequencing of RHD and/or RHCE alleles, additional RH alleles are likely to be identified and the allele assignments made based on medium resolution genotyping methods may need to be updated. A Punnett square is generated for each patient for whom RH allele selected units are requested, based on the RH alleles they carry. Though a Punnett square could be generated with all known RHD and RHCE alleles, those that are Tier 1, 2 and 3 and for which there are donors in the American Rare Donor Program are typically included. This approach could be modified based on the population and the RH characterization of rare donors.
The use of a Punnett square allows donors with the same or similar alleles to be assigned to Tiers.
Table 9 defines the tiers and how they are assigned. Notably, with RHD, zygosity plays a role as well as an appreciation that some RHD variant alleles such as RHD*DIIIa-CE(4-7)-D do not express RhD but instead express an partial C antigen. RHCE alleles “drive” allele selection since matching is typically performed due to anti-e, -e-like, -hrB, -hrS or -Hr antibodies. Importantly patients who carry two different RHCE alleles generally have more potentially compatible donors since the list of donors would include donors carrying the same combination of alleles as the patient (Tier 1) as well as those homozygous for either allele (Tier 2) or those carrying different alleles predicted to encode a similar phenotype.
Table 9
| Tier | RHCE | RHD |
|---|---|---|
| 1 | Probable alleles of the donor are the same as the probable alleles of the patient | Probable alleles of the donor are the same as the probable alleles of the patient, or the donor does not express RhD antigen |
| 2 | The donor is homozygous for one of the probable alleles of the patient | The donor is hemizygous or homozygous for one of the probable alleles of the patient |
| 3 | The donor carries one or two alleles that are not the same but have a similar phenotype as the alleles of the patient | The donor may carry an RHD allele predicted to express a “benign” variant while the patient is predicted to express a normal RhD |
| 4 | The donor carries one or two alleles that are not the same and have a different phenotype than the alleles of the patient. This could be due to gain of low prevalence antigen(s) such as V or VS | The donor carries one or two alleles that are not the same and have a different phenotype than the alleles of the patient. This could be due to gain of low prevalence antigen(s) such as Go(a) or DAK |
A clinical case example of RH allele selection
The ARDP received a request for blood from a blood center outside of the US for an 18-year old female with sickle cell disease. The patient was A positive with anti-C, -e, -Wra and an antibody to a high-prevalence antigen in the Rh system. This antibody was found to be anti-hrB. RHCE genotyping predicted that the patient to carry two different RHCE alleles: RHCE*ceVS.03 [also known as RHCE*ce48C,733G,1006T] and RHCE*ceVS.03 [also known as RHCE*ce733G]. RHD genotyping predicted the patient to carry two different RHD alleles: RHD*03N.01 [also known as RHD*DIIIa-CE(4-7)-D] and RHD*01. Based on this information, the patient has a predicted phenotype of D+ partial C+ E- partial e+ partial c+ V+ VS+ hrS+ and hrB+vw/-. The patient’s plasma was non-reactive at PEG IgG-AGT phase with RBCs from two Tier 2 donors carrying RHD*01 (homozygous or hemizygous) and homozygous for RHCE*ce733G. Tables 10-15 illustrates the use of a Punnett Square of RHCE or RHD alleles in RH allele selection of donors based on a patient’s RH alleles, with Table 10 showing the use of a Punnett Square using RHCE common alleles for illustrative purposes only. The RHCE Punnett square in Table 13 and RHD Punnett square in Table 14 are applicable to the clinical case example. Table 16 lists the allele combinations and number of ABO/Rh compatible donors carrying RH alleles predicted to have the same or similar phenotype as the patient. This case was initially presented at ISBT in 2019 (62). A frozen RBC unit from a Tier 2 donor was shipped. The unit was stored and was reported to have been thawed at a later date and transfused to the patient without incident.
Table 10
| RHCE alleles | RHCE*ce | RHCE*Ce | RHCE*cE | RHCE*CE |
|---|---|---|---|---|
| RHCE*ce | Tier 2 | Tier 1 | Tier 4 | Tier 4 |
| RHCE*Ce | Tier 2 | Tier 4 | Tier 4 | |
| RHCE*cE | Tier 4 | Tier 4 | ||
| RHCE*CE | Tier 4 |
The Punnett Square includes the four normal RHCE alleles. The donor homozygous for RHCE*ce is seen in the top left box assigned Tier 2 since they are homozygous for one of the alleles of the patient and are not predicted to express antigens not expressed by the patient. The donor with one RHCE*ce allele and one RHCE*Ce allele is seen in the next box assigned Tier 1 since both the alleles of the donor match both alleles of the patient. The donor homozygous for RHCE*Ce is assigned Tier 2 since they are homozygous for one of the alleles of the patient and are not predicted to express antigens not expressed by the patient. Donors with RHCE*cE or RHCE*CE in any combination are assigned Tier 4 since they would express E antigen that the patient does not; these are included for illustration purposes only. Tables 11,12 show examples of RHCE Punnett squares and Tables 13,14 show examples of RHD Punnett squares for patients carrying variant Rh antigens. Both RHCE and RHD Punnett squares are generated for each patient case and the larger of the Tier number is assigned to the donor. Red Cell units from donors ranked Tier 1 or Tier 2 are preferred over donors ranked Tier 3, and Tier 3 is preferred over Tier 4 units. There is no evidence that Tier 1 RBCs are superior to Tier 2 RBCs. RBC, red blood cell.
Table 11
| RHCE alleles | RHCE*ce733G | RHCE*ce48C,733G | RHCE*ce48C,733G,1006T | RHCE*ce48C | RHCE*ce |
|---|---|---|---|---|---|
| RHCE*ce733G | Tier 1 | Tier 3 | Tier 3 | Tier 4 | Tier 4 |
| RHCE*ce48C,733G | Tier 3 | Tier 3 | Tier 4 | Tier 4 | |
| RHCE*ce48C,733G,1006T | Tier 3 | Tier 4 | Tier 4 | ||
| RHCE*ce48C | Tier 4 | Tier 4 | |||
| RHCE*ce | Tier 4 |
A donor who is homozygous for the RHCE*ce733G allele is assigned to Tier 1 since they have the same alleles as the patient. Donors with other RHCE*ce alleles with hrB- phenotype are expected to be compatible but since not identical are assigned Tier 3. Neither the consensus allele RHCE*ce nor the common variant RHCE*ce48C lack the high-prevalence antigen hrB and therefore are not expected to be compatible with the patient who has made allo-anti-e or-hrB, therefore they are assigned Tier 4.
Table 12
| RHCE alleles | RHCE*ce48C,733G,1006T | RHCE*ce48C,733G | RHCE*ce733G | RHCE*ce48C | RHCE*ce |
|---|---|---|---|---|---|
| RHCE*ce48C,733G,1006T | Tier 1 | Tier 3 | Tier 3 | Tier 4 | Tier 4 |
| RHCE*ce48C,733G | Tier 3 | Tier 3 | Tier 4 | Tier 4 | |
| RHCE*ce733G | Tier 3 | Tier 4 | Tier 4 | ||
| RHCE*ce48C | Tier 4 | Tier 4 | |||
| RHCE*ce | Tier 4 |
The Punnett square includes the allele carried by the patient as well as the RHCE*ce48C,733G allele that also lacks hrB and the RHCE*ce733G allele with hrB+vw/- phenotype. Since these two alleles are not identical and also express V, they are assigned Tier 3. Alleles that would not be considered include RHCE*ce or RHCE*ce48C because they do not lack the hrB antigen; these are assigned Tier 4.
Table 13
| RHCE alleles | RHCE*ce48C,733G,1006T | RHCE*ce733G | RHCE*ce48C,733G | RHCE*ce48C | RHCE*ce |
|---|---|---|---|---|---|
| RHCE*ce48C,733G,1006T | Tier 2 | Tier 1 | Tier 3 | Tier 4 | Tier 4 |
| RHCE*ce733G | Tier 2 | Tier 3 | Tier 4 | Tier 4 | |
| RHCE*ce48C,733G | Tier 3 | Tier 4 | Tier 4 | ||
| RHCE*ce48C | Tier 4 | Tier 4 | |||
| RHCE*ce | Tier 4 |
The Punnett square includes the allele carried by the patient as well as the RHCE*ce48C,733G allele that also lacks hrB and the RHCE*ce733G allele with hrB+vw/- phenotype. Since these two alleles are not identical and also express V, they are assigned Tier 3. Alleles that would not be considered include RHCE*ce or RHCE*ce48C because they do not lack the hrB antigen; these are assigned Tier 4 for illustrative purposes.
Table 14
| RHD alleles | RHD*DIIIa-CE(4-7)-D | RHD*01 | RHD*01N.01 | RHD*08N.01 | RHD*DAU0 | RHD*weak D type 4.0 |
|---|---|---|---|---|---|---|
| RHD*DIIIa-CE(4-7)-D | Tier 2 | Tier 1 | Tier 2 | Tier 2 | Tier 3 | Tier 3 |
| RHD*01 | Tier 2 | Tier 2 | Tier 2 | Tier 3 | Tier 3 | |
| RHD*01N.01 | Tier 1 | Tier 1 | Tier 3 | Tier 3 | ||
| RHD*08N.01 | Tier 1 | Tier 3 | Tier 3 | |||
| RHD*DAU0 | Tier 3 | Tier 3 | ||||
| RHD*weak D type 4.0 | Tier 3 |
This Punnett square includes donors who carry one or two RHD*DIIIa-CE(4-7)-D alleles predicted to express an altered C antigen, or two conventional RHD*01 alleles, or one of each. The donor who is homozygous for one of the two distinct alleles carried by the patient are ranked Tier 2, while donors with both alleles are ranked Tier 1. Also included as options are the non-functional RHD*08N.01 or deleted RHD*01N.01, both of which have a D- phenotype and would be ranked Tier 1. Also included are alleles that encode what are considered “benign variants” RHD*DAU0 and RHD*Weak D Type 4.0; these are ranked Tier 3. When RHD and RHCE tiers are determined for a donor, that donor’s tier rank is the higher value. For example, if the donor carries identical RHD alleles (Tier 1) but is homozygous for one RHCE allele in a patient with two distinct alleles (Tier 2), the overall Tier for that donor would be Tier 2.
Table 15
| RHD alleles | RHD*DIIIa-CE(4-7)-D | RHD*01N.01 | RHD*08N.01 |
|---|---|---|---|
| RHD*DIIIa-CE(4-7)-D | Tier 1 | Tier 1 | Tier 1 |
| RHD*01N.01 | Tier 1 | Tier 1 | |
| RHD*08N.01 | Tier 1 |
This Punnett square includes donors who carry one or two RHD*DIIIa-CE(4-7)-D alleles predicted to express an altered C antigen. No donors with conventional RHD*01 or benign variant alleles such as RHD*DAU0 or RHD alleles encoding partial D antigens are included since these would be D+ and therefore not compatible with the patient. There are fewer options for patients who do not express RhD antigen or who carry a hybrid gene that expresses altered C antigen. Donors that have non-functional RHD allele(s) (e.g., RHD*08N.01) or have RHD gene(s) deleted (RHD*01N.01), though they carry different alleles than the patient, are ranked Tier 1 since they do not encode a protein product nor any antigens. Any donors with RHD alleles that express normal, weak or partial RhD antigens would be ranked Tier 4.
Table 16
| RH Haplotype | Donors | |||
|---|---|---|---|---|
| Probable RHD alleles | Probable RHCE alleles | N | Tier | |
| RHD*01/RHD-CE(4-7)-D | RHCE*ce733G/RHCE*ce48C,733G,1006T | 102 | 1 | |
| RHD*01 (homoz/hemiz) | RHCE*ce733G/RHCE*ce48C,733G,1006T | 4 | 2 | |
| RHD-CE(4-7)-D (homoz/hemiz) | RHCE*ce48C,733G,1006T homozygous | 36 | 2 | |
| RHD*01/RHD-CE(4-7)-D | RHCE*ce48C,733G,1006T homozygous | 7 | 2 | |
| RHD*01 (homoz/hemiz) | RHCE*ce48C,733G,1006T/RHCE*ce733G | 6 | 2 | |
| RHD-CE(4-7)-D (homoz/hemiz) | RHCE*ce48C,733G,1006T/RHCE*ce733G | 5 | 2 | |
| RHD*01 (homoz/hemiz) | RHCE*ce733G homozygous | 88 | 2 | |
| RHD*01/RHD-CE(4-7)-D | RHCE*ce733G homozygous | 1 | 2 | |
| RHD*01N.01 homozygous | RHCE*ce733G homozygous | 1 | 2 | |
| RHD not tested | RHCE*ce733G homozygous | 357 | 2 | |
| RHD*01 (homoz/hemiz) | RHCE*ce48C,733G/RHCE*ce733G | 2 | 3 | |
| RHD*01 (homoz/hemiz) | RHCE*ce48C,733G homozygous | 7 | 3 | |
| RHD*01 (homoz/hemiz) | RHCE*ce733G/RHCE*ce48C,733G,1006T | 4 | 3 | |
| RHD*01/RHD-CE(4-7)-D | RHCE*ce48C,733G/RHCE*ce48C,733G,1006T | 3 | 3 | |
| RHD*01/ RHD*DAU0 | RHCE*ce733G/RHCE*ce48C,733G | 2 | 3 | |
| RHD*01/ RHD*DAU0 | RHCE*ce48C,733G homozygous | 1 | 3 | |
| RHD*01/ RHD*DAU0 | RHCE*ce733G homozygous | 1 | 3 | |
| RHD not tested | RHCE*ce48C,733G homozygous | 1 | 3 | |
| RHD not tested | RHCE*ce48C,733G/RHCE*ce48C,733G,1006T | 12 | 3 | |
| RHD not tested | RHCE*ce733G/RHCE*ce48C,733G,1006T | 50 | 3 | |
| RHD not tested | RHCE*ce733G/RHCE*ce48C,733G | 5 | 3 | |
homoz/hemiz, homozygous or hemizygous.
A Review of ARDP activities involving RH allele selection
A review of ARDP cases from the year 2020 involving RH allele selected donor units showed that of the 1,045 requests for rare blood, 126 (12%) requests involving 53 patients were for RH allele selected units. Nearly half (26 of 53) of these patients carried the RHCE*ce733G allele, with 11 (42%) reported to have anti-hrB and 5 (19%) allo-anti-e or allo-anti-e-like antibody with 10 (38%) having warm reactive autoantibodies, 3 (12%) having cold reactive autoantibodies and 11 (42%) have multiple alloantibodies. At that time, the ARDP database included 1,979 donors that carried the RHCE*ce733G allele with 982 (50%) being homozygous. Of the requests for allele-selected units for patients with an RHCE*ce733G allele, 6 (8%) were unable to be filled; these 6 requests required the units to also be negative for C, E, K, S, Fya with three requiring Fy(b-), two requiring N- and two requiring Jk(b-) units. This analysis illustrates that the ability to fill RH allele selected unit requests is impacted not only by the number of donors with RH allele information but also by the presence of multiple alloantibodies in the patient. ABO type compatibility also plays a role in the number of donors whose units could fill such requests (65).
Challenges to RH allele selection: ambiguous phenotypes
A factor that complicates using RH allele selected donor units for Rh alloimmunized patients with partial Rh antigens is ambiguous phenotypes. These include the hrB+vw/-phenotype assigned to red cells carrying the RHCE*ce733G allele. This phenotype is confusing to clinicians who need to be able to interpret alloimmunization risk, to manufacturers of genotyping kits that include algorithms to assign predicted phenotypes as well as to reference laboratories asked to provide transfusion recommendations based on molecular reports. Ambiguity also lies in the alloimmunization risk for anti-D in patients carrying the RHD*DAU0 and RHD*weak D type 4.0 alleles (52). This is also true of the RHCE*ce48C allele found in more than 10% of African American blood donors (66) for which ISBT Working Party on Red Cell Immunogenetics and Blood Group Terminology (1) assigns a phenotype of e+weak yet allo-anti-e has been reported in individuals homozygous for this allele (67). Finally, transfusion medicine professionals are trained to provide antigen negative blood products for alloimmunized patients. However, an RH allele selected donor unit may type positive for one or more of the Rh antigens for which a patient may have antibodies, yet the unit would be predicted to be compatible since both the donor and the patient express the same or similar partial antigen(s). Finally, neither hospital computer systems nor blood establishment computer systems are likely equipped to store RH allele information such that it can be used by transfusion medicine professionals in the selection of blood products. Due to the many RHCE alleles expressing partial e antigens, some with incomplete information regarding clinically relevant Rh antigen phenotype classifications, it would be beneficial for the transfusion medicine community to classify this group of antigens eVAR; this approach would be similar to that taken with UVAR (68) and would allow databases to use this “new” antigen to overcome limitations of the current binary status (e+ or e−) (69).
Discussion/summary
Though there are limitations to our current understanding the genetics of the RH blood group system and the implications of the many variant alleles and resulting antigens on alloimmunization risk, the benefits of using RH allele information for clinical care are clear. Multiple studies, many of which were reviewed here, have demonstrated clear benefit to the use of RHD genotyping to provide allele information that can be used to assess alloimmunization risk. The value of this methodology is most obvious in women of child-bearing age to determine who would benefit from Rh immune prophylaxis and who does not require it. Also, in patients with variant RHCE alleles, selection of RBC units from donors with the same or similar alleles holds great promise to deliver personalized transfusion medicine, especially in patient populations where chronic transfusion is associated with high rates of alloimmunization. Whereas the use of RH allele selected donor units is currently limited to patients with Rh alloantibodies, if donor centers can attract and retain donors of African descent and can identify those with variant RH alleles lacking high prevalence antigens, there would be the potential to use this approach prophylactically.
Both in reducing risk of allo-anti-D formation and in identification and selection of compatible blood products for patients who have made antibodies to high prevalence antigens, molecular methods can be instrumental in developing a personalized approach to patient care.
Acknowledgments
I thank Sandra T Nance, MS, MT(ASCP) SBB, retired, for critical review of the manuscript.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Lilian Castilho) for the series “Serology and Molecular Biology of the Rh System” published in Annals of Blood. The article has undergone external peer review.
Reporting Checklist: The author has completed the Narrative Review reporting checklist. Available at https://aob.amegroups.com/article/view/10.21037/aob-22-6/rc
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://aob.amegroups.com/article/view/10.21037/aob-22-6/coif). The series “Serology and Molecular Biology of the Rh System” was commissioned by the editorial office without any funding or sponsorship. MAK reports that she is an employee of American Red Cross. She also reports support for travel to Annual Meetings (Boston and Chicago) from American Society Clinical Pathology and she is the secretary of ISBT Working Party and South Central Association of Blood Banks. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Available online: https://www.isbtweb.org/working-parties/red-cell-immunogenetics-and-blood-group-terminology/
- Cartron JP, Rouillac C, Le Van Kim C, et al. Tentative model for the mapping of D epitopes on the RhD polypeptide. Transfus Clin Biol 1996;3:497-503. [Crossref] [PubMed]
- Flegel WA, Wagner FF. RHD epitope density profiles of RHD variant red cells analyzed by flow cytometry. Transfus Clin Biol 1996;3:429-31. [Crossref] [PubMed]
- STRATTON F.. A new Rh allelomorph. Nature 1946;158:25. [Crossref] [PubMed]
- Scientific Committee of the Joint Blood Council & Standards Committee of the American Association of Blood Banks. Standards for a Blood Transfusion Service. Washington, DC: Joint Blood Council Inc., 1958.
- Tippett P, Sanger R. Observations on subdivisions of the Rh antigen D. Vox Sang 1962;7:9-13. [Crossref] [PubMed]
- Agre PC, Davies DM, Issitt PD, et al. A proposal to standardize terminology for weak D antigen. Transfusion 1992;32:86-7. [Crossref] [PubMed]
- Daniels G. Variants of RhD--current testing and clinical consequences. Br J Haematol 2013;161:461-70. [Crossref] [PubMed]
- Sandler SG, Roseff SD, Domen RE, et al. Policies and procedures related to testing for weak D phenotypes and administration of Rh immune globulin: results and recommendations related to supplemental questions in the Comprehensive Transfusion Medicine survey of the College of American Pathologists. Arch Pathol Lab Med 2014;138:620-5. [Crossref] [PubMed]
- Sandler SG, Flegel WA, Westhoff CM, et al. It’s time to phase in RHD genotyping for patients with a serologic weak D phenotype. College of American Pathologists Transfusion Medicine Resource Committee Work Group. Transfusion 2015;55:680-9. [Crossref] [PubMed]
- Joint Statement on Phasing-In RHD Genotyping for Pregnant Women and Other Females of Childbearing Potential with a Serologic Weak D Phenotype. AABB Advocacy Statement 2015. Available online: https://www.aabb.org/docs/default-source/default-document-library/positions/statement150722.pdf?sfvrsn=f4f44c30_6
- Kacker S, Vassallo R, Keller MA, et al. Financial implications of RHD genotyping of pregnant women with a serologic weak D phenotype. Transfusion 2015;55:2095-103. [Crossref] [PubMed]
- Sandler SG, Chen LN, Flegel WA. Serological weak D phenotypes: a review and guidance for interpreting the RhD blood type using the RHD genotype. Br J Haematol 2017;179:10-9. [Crossref] [PubMed]
- Denomme GA, Wagner FF, Fernandes BJ, et al. Partial D, weak D types, and novel RHD alleles among 33,864 multiethnic patients: implications for anti-D alloimmunization and prevention. Transfusion 2005;45:1554-60. [Crossref] [PubMed]
- Noizat-Pirenne F, Verdier M, Lejealle A, et al. Weak D phenotypes and transfusion safety: where do we stand in daily practice? Transfusion 2007;47:1616-20. [Crossref] [PubMed]
- Denomme GA, Dake LR, Vilensky D, et al. Rh discrepancies caused by variable reactivity of partial and weak D types with different serologic techniques. Transfusion 2008;48:473-8. [Crossref] [PubMed]
- Luo X, Keller MA, James I, et al. Strategies to identify candidates for D variant genotyping. Blood Transfus 2018;16:293-301. [Crossref] [PubMed]
- Horn TN, Keller J, Keller MA, et al. Identifying obstetrics patients in whom RHD genotyping can be used to assess risk of D alloimmunization. Immunohematology 2020;36:146-51.
- Hudgins JP, Matsushita C, Tuma CW, et al. Identification of RHD allelic variants discovered by atypical typing results on the NEO/Echo platforms. Immunohematology 2021;37:165-70. [Crossref] [PubMed]
- Keller M, Lindquist M, Smith A, et al. Analysis of RHD alleles in patients in the USA. Vox Sanguinis 2021;116:38.
- Wang D, Lane C, Quillen K. Prevalence of RhD variants, confirmed by molecular genotyping, in a multiethnic prenatal population. Am J Clin Pathol 2010;134:438-42. [Crossref] [PubMed]
- Campos FC, Mota MA, Aravechia MG, et al. Variant RHD Types in Brazilians With Discrepancies in RhD Typing. J Clin Lab Anal 2016;30:845-8. [Crossref] [PubMed]
- Bub CB, Aravechia MG, Costa TH, et al. RHD alleles among pregnant women with serologic discrepant weak D phenotypes from a multiethnic population and risk of alloimmunization. J Clin Lab Anal 2018;32:e22221. [Crossref] [PubMed]
- Dezan MR, Oliveira VB, Gomes ÇN, et al. High frequency of variant RHD genotypes among donors and patients of mixed origin with serologic weak-D phenotype. J Clin Lab Anal 2018;32:e22596. [Crossref] [PubMed]
- Oodi A, Daneshvar Z, Goudarzi S, et al. RHD genotyping of serological weak D phenotypes in the Iranian blood donors and patients. Transfus Apher Sci 2020;59:102870. [Crossref] [PubMed]
- Thongbut J, Laengsri V, Raud L, et al. Nation-wide investigation of RHD variants in Thai blood donors: Impact for molecular diagnostics. Transfusion 2021;61:931-8. [Crossref] [PubMed]
- Chung YN, Kim TY, Yu H, et al. Molecular basis of serological weak D phenotypes and RhD typing discrepancies identified in the Korean population. Blood Transfus 2021;19:327-34. [Crossref] [PubMed]
- Bakry RM, Nasreldin E, Hassaballa AE, et al. Evaluation of molecular typing and serological methods in solving discrepant results of weak and partial D (Rh) in South Egypt. Asian J Transfus Sci 2019;13:110-4. [Crossref] [PubMed]
- Yan L, Wu J, Zhu F, et al. Molecular basis of D variants in Chinese persons. Transfusion 2007;47:471-7. [Crossref] [PubMed]
- Ouchari M, Srivastava K, Romdhane H, et al. Transfusion strategy for weak D Type 4.0 based on RHD alleles and RH haplotypes in Tunisia. Transfusion 2018;58:306-12. [Crossref] [PubMed]
- Standard for Blood Banks and Transfusion Services, 32nd Edition, AABB 2020.
- Wagner FF, Kasulke D, Kerowgan M, et al. Frequencies of the blood groups ABO, Rhesus, D category VI, Kell, and of clinically relevant high-frequency antigens in south-western Germany. Infusionsther Transfusionsmed 1995;22:285-90. [Crossref] [PubMed]
- Flegel WA, Denomme GA, Queenan JT, et al. It’s time to phase out "serologic weak D phenotype" and resolve D types with RHD genotyping including weak D type 4. Transfusion 2020;60:855-9. [Crossref] [PubMed]
- Tahhan HR, Holbrook CT, Braddy LR, et al. Antigen-matched donor blood in the transfusion management of patients with sickle cell disease. Transfusion 1994;34:562-9. [Crossref] [PubMed]
- Vichinsky EP, Luban NL, Wright E, et al. Prospective RBC phenotype matching in a stroke-prevention trial in sickle cell anemia: a multicenter transfusion trial. Transfusion 2001;41:1086-92. [Crossref] [PubMed]
- Castro O, Sandler SG, Houston-Yu P, et al. Predicting the effect of transfusing only phenotype-matched RBCs to patients with sickle cell disease: theoretical and practical implications. Transfusion 2002;42:684-90. [Crossref] [PubMed]
- Lasalle-Williams M, Nuss R, Le T, et al. Extended red blood cell antigen matching for transfusions in sickle cell disease: a review of a 14-year experience from a single center (CME). Transfusion 2011;51:1732-9. [Crossref] [PubMed]
- Ribeiro KR, Guarnieri MH, da Costa DC, et al. DNA array analysis for red blood cell antigens facilitates the transfusion support with antigen-matched blood in patients with sickle cell disease. Vox Sang 2009;97:147-52. [Crossref] [PubMed]
- Wilkinson K, Harris S, Gaur P, et al. Molecular blood typing augments serologic testing and allows for enhanced matching of red blood cells for transfusion in patients with sickle cell disease. Transfusion 2012;52:381-8. [Crossref] [PubMed]
- Davis BA, Allard S, Qureshi A, et al. Guidelines on red cell transfusion in sickle cell disease. Part I: principles and laboratory aspects. Br J Haematol 2017;176:179-91. [Crossref] [PubMed]
- Chou ST, Alsawas M, Fasano RM, et al. American Society of Hematology 2020 guidelines for sickle cell disease: transfusion support. Blood Adv 2020;4:327-55. [Crossref] [PubMed]
- Klapper E, Zhang Y, Figueroa P, et al. Toward extended phenotype matching: a new operational paradigm for the transfusion service. Transfusion 2010;50:536-46. [Crossref] [PubMed]
- Castilho L, Dinardo CL. Optimized Antigen-Matched in Sickle Cell Disease Patients: Chances and Challenges in Molecular Times - the Brazilian Way. Transfus Med Hemother 2018;45:258-62. [Crossref] [PubMed]
- Sesok-Pizzini DA, Friedman DF, Smith-Whitley K, et al. Transfusion support of patients with sickle cell disease at the Children’s Hospital of Philadelphia. Immunohematology 2006;22:121-5.
- Pham BN, Peyrard T, Juszczak G, et al. Heterogeneous molecular background of the weak C, VS+, hr B-, Hr B- phenotype in black persons. Transfusion 2009;49:495-504. [Crossref] [PubMed]
- Chou ST, Jackson T, Vege S, et al. High prevalence of red blood cell alloimmunization in sickle cell disease despite transfusion from Rh-matched minority donors. Blood 2013;122:1062-71. [Crossref] [PubMed]
- Dinardo CL, Kelly S, Dezan MR, et al. Diversity of RH and transfusion support in Brazilian sickle cell disease patients with unexplained Rh antibodies. Transfusion 2019;59:3228-35. [Crossref] [PubMed]
- Faas BH, Beckers EA, Wildoer P, et al. Molecular background of VS and weak C expression in blacks. Transfusion 1997;37:38-44. [Crossref] [PubMed]
- Nance ST, Keller MA. Comments on: molecular matching of red blood cells is superior to serological matching in sickle cell disease patients. Rev Bras Hematol Hemoter 2013;35:9-11. [Crossref] [PubMed]
- Fasano RM, Sullivan HC, Bray RA, et al. Genotyping Applications for Transplantation and Transfusion Management: The Emory Experience. Arch Pathol Lab Med 2017;141:329-40. [Crossref] [PubMed]
- Chou ST, Evans P, Vege S, et al. RH genotype matching for transfusion support in sickle cell disease. Blood 2018;132:1198-207. [Crossref] [PubMed]
- Floch A, Pirenne F, Barrault A, et al. Insights into anti-D formation in carriers of RhD variants through studies of 3D intraprotein interactions. Transfusion 2021;61:1286-301. [Crossref] [PubMed]
- Karafin MS, Field JJ, Gottschall JL, et al. Barriers to using molecularly typed minority red blood cell donors in support of chronically transfused adult patients with sickle cell disease. Transfusion 2015;55:1399-406. [Crossref] [PubMed]
- Crowley JA, Horn T, Keller MA. RHCE Characterization requires both commercial genotyping arrays and lab-developed tests. Transfusion 2013;53:172A.
- Montemayor C, Brunker PAR, Keller MA. Banking with precision: transfusion medicine as a potential universal application in clinical genomics. Curr Opin Hematol 2019;26:480-7. [Crossref] [PubMed]
- Meny GM, Flickinger C, Marcucci C. The American Rare Donor Program. J Crit Care 2013;28:110.e9-110.e18. [Crossref] [PubMed]
- Castilho L, Flickinger C, Horn T, et al. The Importance of molecular testing to Identify and provide compatible donor units for patients with anti-hrB and anti-hrS. Vox Sanguinis 2011;101:269.
- Keller M, Horn T, Crowley JA, et al. Genotype Compatibility Tables for Matching Patients and Donors for Rh Variants. Transfusion 2013;53:174A.
- Keller MA, Horn T, Fludd DR, et al. RH Allele Matching for Rh Alloimmunized Patients: Personalized Medicine to the Nth Degree. Transfusion 2019;59:143A.
- Keller MA, Kavitsky V, Maurer JL, et al. RHCE*ce733G, Alloimmunization and Meeting the Blood Needs of Alloimmunized Patients. Transfusion 2021;61:145A.
- Bhoj V, Blair P, Friedman D, et al. An Anti-hrB-like Antibody in a Sickle Cell Patient with a Variant e Confirmed by Molecular Testing. Transfusion 2013;53:121A.
- Keller MA, Mansfield P, Maurer J, et al. Identifying compatible blood for Rh alloimmunized patients: An international case. Vox Sanguinis 2019;114:85.
- Keller MA, Keller J, Thomas D, et al. Genotyping Panel designed to identify blood donors lacking high prevalence Rh antigens. Vox Sanguinis 2018;113:252.
- Thomas DE, Keller MA. High Throughput RH Genotyping of More than 500 African American Blood Donors. Transfusion 2021;61:132A.
- Maurer JL, Nance ST, Kavitsky V, et al. High Prevalence Antigen Negative Unit Requests: What If It Includes Being O Negative? Transfusion 2021;60:159A.
- Keller MA, Crowley J, Horn T, et al. Characterization of RH locus in African American Blood Donors. Transfusion 2013;53:28A.
- Lapadat R, Meyer E, Josephson C, et al. Exon Sequencing Improves Red Blood Cell Phenotype Matching in a Patient Homozygous for the RHCE*ce48C Allele. 2015. Transfusion 2015;55:152A.
- Storry JR, Reid ME, Fetics S, et al. Mutations in GYPB exon 5 drive the S–s–U+var phenotype in persons of African descent: implications for transfusion. 2003. Transfusion 2003;43:1738-47. [Crossref] [PubMed]
- Nance SN, Mansfield P. Use of eVAR and anti-eVAR as interim terms. Immunohematology 2022; In press. [Crossref]
Cite this article as: Keller MA. RH genetic variation and the impact for typing and personalized transfusion strategies: a narrative review. Ann Blood 2023;8:18.


