
Hybrids and microconversions in RH genes: investigation and implication in transfusion therapy
Introduction
In the early nineties and more than 50 years after the identification of the D antigen, the molecular basis of the RH blood group system (ISBT #004) was deciphered. It did not take a long time thereafter before genetic variants associated with altered antigen expression were found in the genes. And still 30 years later, novel variations are reported on a regular basis in the scientific literature, notably by taking advantage of the contemporary technologies coming up in the market place, accounting for hundreds of simple and complex variant alleles. From a clinical point of view, expression of those variant alleles may result in variable expression of RH antigen(s) at both the quantitative and qualitative levels with potential clinical outcomes. In diagnostics, molecular immunohematology has become routine procedure in reference laboratories for predicting phenotype and guiding transfusion practice as a function of the risk of alloimmunization. The basic knowledge of RH molecular genetics is thus the key point of such a strategy.
In the RH genes, variant alleles mostly involve single nucleotide variant (SNVs) and/or structural variants (SVs), the latter basically altering large genomic sequences. A specific feature of RH molecular genetics is the transfer of genetic materials from one gene to another (= gene conversion), thereby creating ‘hybrid genes’ (that will be discussed in the next section). In this context, this review aims (I) to provide a global and nonexhaustive overview of this latter category of alleles from a genomic point of view, (II) to report their association with antigen expression, (III) to describe their frequency in various populations and, finally, (IV) to discuss their identification and relevance in daily practice in diagnostics and for the management of transfusion.
Molecular genetics of RH hybrid genes: a genomic perspective
The basics of RH molecular genetics
The paralogous RHD and RHCE genes involved in the RH blood group system were identified in the early nineties. Both genes are composed by ten exons and are separated by the TMEM50A gene within the RH locus on the short arm of human chromosome 1 (1-6). RHD is the duplicated product of the ancestral RHCE gene (7). RHD and RHCE encode the multipass transmembrane RhD and RhCE proteins, respectively. The former carries the D (or RH1) antigen, while the latter harbours the antithetical C/c (RH2/RH4) and E/e (RH3/RH5) antigens (8,9).
The generation of RH hybrid genes is directly linked to the structure of the RH locus
In 1994, soon after the description of the genes, Mouro and colleagues investigated the molecular basis of ‘DVI category phenotype’ in blood donors by Southern-blotting and reverse transcription-polymerase chain reaction (RT-PCR) analysis (10). For the first time, they described a gene conversion event encompassing exons 4, 5, and 6 within the RHD gene, i.e., RHD-CE(4-6)-D, which is currently referred to as the DVI.2 (or RHD*06.02) allele. At the end of the year, the same team reported RHCE-D-CE hybrid alleles by probe hybridization and transcript analysis in samples presenting with the rare Dc– and DCW– phenotypes (11). Both reports are milestones towards the identification and characterization of hybrid genes.
From a genomic point of view, homology, physical proximity and opposite orientation of the RH genes, as well as the homologous 5'- and 3'-Rhesus boxes at both sides of RHD, are actually critical and favourable factors supporting potential genetic rearrangements, such as unequal crossing-over—as illustrated by the deletion of the whole RHD locus generating the common D-negative RHD*01N.01 allele –, and gene conversion (12). Typically, the latter mutational event involves the transfer of genetic materials from the ‘donor’ gene that replace the paralogous sequence within the ‘recipient’ gene (13). The novel alleles created by this mechanism are referred to as RHD-CE-D (from RHCE to RHD) and RHCE-D-CE (from RHD to RHCE) ‘hybrid alleles’—or ‘hybrid genes’—and exhibit both RHD and RHCE molecular specificity. The size of the genomic transfer is highly variable, ranging from the microconversion of a single nucleotide (e.g., RHD*DFV, *DCS2, and *DUC2) up to large genomic blocks, composed by exons and introns, spanning several dozen kilobases (e.g., RHD*01N.03, *01N.04, and *01N.05) (13-18).
The relationship between RH hybrid genes and antigen expression
Although the global genomic architecture of those variant alleles is not modified when compared to a ‘normal’ RH gene, i.e., 10 ‘RH’ exons separated alternatively by nine ‘RH’ introns, substitution of gene-specific coding DNA sequence by its counterpart changes the sequence of the translated ‘hybrid’ protein, notably by introducing missense variants from one gene to another, namely ‘templated’ variants. For example, variant alleles from the DIVa and weak D type 4 clusters carrying the c.455A>C and c.667T>G microconversions in RHD (19,20), respectively, may be considered typically as ‘hybrid alleles’. Change in the amino acid sequence has thereby the potential to alter quantitatively but also, and more importantly, qualitatively the expression of the polymorphic D antigen (if occurring at the RHD locus) or the C, c, E, and/or e antigens (if occurring at the RHCE locus). Indeed, those alterations directly disrupt the structure and/or the primary sequence of the protein product. The biosynthesized hybrid protein thus carries an incomplete number of epitopes, as revealed by the patterns of reaction obtained with various monoclonal antibodies (21-25), resulting in a so-called ‘partial phenotype’ (e.g., partial D, partial c, etc.). As an example, in the model of 30 epitopes defining the D antigen, the vast majority is lacking in DVI RBCs, while most of them are present in DVa RBCs (26,27). Consequently, from a clinical point of view, those individuals exhibiting a partial phenotype may produce an alloantibody if exposed to the corresponding normal antigen (= alloimmunization) [see (28) for a recent review on RH gene variants and antibody production]. The nature and specificity of alloantibodies have a direct impact on the severity of the hemolytic reaction that may occur potentially if exposed again to the same antigen(s) in a transfusion and/or obstetrical context(s).
Finally, hybrid proteins may not express high-frequency antigens (HFAs), which are thus absent from the RBC membrane, and/or create novel sequences and structural motifs carrying low-frequency (or rare) antigens (LFAs), as illustrated by the following examples throughout the paper. Overall, the study of the molecular structure of hybrid genes and its related expression has gained much attention with an evident clinical interest, and has definitely contributed to the global understanding of the complex RH blood group system.
Expression of RH hybrid genes: an ‘antigen’ perspective
RH hybrid genes and variant phenotype
After the initial discoveries made by the French team, several other hybrid genes associated with the expression of the so-called DIIIc, DIVa, DIVb, DV and DFR partial phenotypes were characterized by the same group and others (29-31). These findings suggested that gene rearrangement is a redundant mutational mechanism resulting in RH variant phenotype and illustrated the growing complexity of RH molecular genetics at that time. Interestingly from a clinical point of view and for diagnostics, the resolution of hybrid variant alleles helped for defining the molecular basis of several LFAs, including Goa (RH30), DW (RH23), FPTT (RH50), RH32, and BARC (RH52) that are further discussed below (Table 1).
Table 1
| Antigen | Clinical relevance (references) |
|---|---|
| VS (RH20) | Mild HDFN (32) |
| DW (RH23) | Severe HDFN (33) |
| Goa (RH30) | Severe HDFN (34-36), DHTR (37) |
| RH32 | Severe HDFN (38-40) |
| Evans (RH37) | HDFN (41) |
| RH42 | Mild HDFN (42) |
| STEM (RH49) | Mild HDFN (43) |
| DAK (RH54) | Severe HDFN (40) |
No clinical significance has been yet recorded for antibodies directed against the RH33, RH43, FPTT (RH50), BARC (RH52) and CENR (RH56) LFAs. LFA, low-frequency antigen; HDFN, hemolytic disease of the fetus and newborn; DHTR, delayed hemolytic transfusion reaction.
DIV alleles and Goa antigen expression
DIVa and DIVb RBCs can be distinguished by their differential expression of Goa (44,45); the former being Go(a+), while the latter are Go(a–). Rouillac and colleagues proposed a tentative structure of the RHD*DIVa allele associated with Goa antigen expression, consisting of two microconversions in exons 3 and 7, as well as an untemplated missense SNV in exon 2, resulting in three amino acid substitutions at the protein level: p.Asn152Thr, p.Asp350His and p.Leu62Phe, respectively (29). This allele has never been confirmed formally since, but a similar structure completed by the untemplated p.Ala137Val substitution, defining the likely RHD*DIVa (or *04.01) allele presenting with the same phenotype, was later described (46) (Figure 1). In contrast, the RHD*DIVb (or *04.06) allele expressed in the Go(a–) DIVb RBCs consists in a gene conversion from a partial exon 7 (from c.1048) up to exon 9, i.e., RHD-CE(7:1048-9)-D, accounting for a total of four templated missense changes in the hybrid protein (29).

Three additional DIV hybrid alleles have been characterized since (47-49), all of them being typed Go(a–) (Figure 1). On the basis of these observations, the templated p.Asp350His change in RhD appears to be required, but not sufficient, to the expression of Goa (20).
DV alleles and DW antigen expression
First, RHD*DV.1 and *DV.2 (29), and later RHD*DV.4 (50,51), *DV.8 (50,51), *DAU5 (16,18), and *DV.10 (52), which all involve p.Glu233Gln in all or part of RHCE exon 5 (Figure 2), were shown to express DW (55). Interestingly, among RHD*DV.4, *DV.5, and *DFV that differ from the reference sequence by a single missense variant (i.e., p.Glu233Gln, p.Glu233Lys, and p.Phe223Val, respectively), only the former expresses DW (15,18,50,51) (Figure 2). Also, RHD*DV.2 and *DBS1 alleles both consist in a RHD-CE(5)-D hybrid gene, but differ by the single polymorphism encoding E and e, respectively, i.e., p.Ala226 and p.Pro226 (29,53,54). RBCs from these allele carriers are respectively DW+ and DW– (Figure 2). These results indicate that both p.Gln233, induced by a microconversion, and p.Ala226, which is specific to both RHD and RHCE carrying a ‘e’ allele, are critical to the expression of DW.
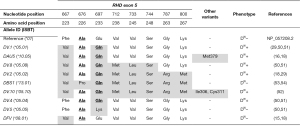
Hybrid alleles and FPTT antigen expression
Expression of FPTT in DFR RBCs was shown in 1988 (56,57). The year after, Rouillac and colleagues reported the structure of the RHD*DFR1 (or *17.01) allele, which consists in the conversion of a discrete region of RHD by the paralogous sequence of RHCE in exon 4, i.e., RHD-CE(4:505-514)-D, thereby introducing the three p.Met169Leu, p.Met170Arg, and p.Ile172Phe missense variants in RhD (29). Other DFR hybrid alleles have been reported since, all of them sharing the first two amino acid substitutions (58-60) (Figure 3).

FPTT-positive expression has also been reported in RBCs expressing the DHAR (or ceHAR) haplotype in the rare R0Har RBCs, and the RHCE*CeVa (or *02.04) allele, which are both composed by a RHCE-D(5)-CE hybrid gene, but on a different allelic background (61,62) (Figure 3). DHAR haplotype is defined by the single aforementioned hybrid allele and, although composed by a single ‘RHD-like exon’, expresses a weak partial D antigen with a very limited number of D epitopes in addition to normal c, weak e, and LFA RH33 (67-69). The CeVa allele encodes weak e and RH33, but also partial C (62). As this allele was found in a D-positive individual, it is unknown whether it encodes or not a partial D antigen (Figure 3).
Finally, more than 50 years ago, a report described an Ivorian woman presenting with the so-called DIV(C)– (or DIVa(C)–) phenotype, who experienced a fatal haemolytic disease of the fetus and newborn in her third child (70). Her RBCs were shown to be positive for D (partial), C (very weak), G (RH12), as well as HFAs RH29, Nou (RH44), and Dav (RH47), and LFAs Goa, RH33, Riv (RH45), FPTT; but negative for E and e (56,70-73). The DIVa(C)– haplotype was resolved in 2012 by genomic DNA study and transcript analysis (63). It consists of the RHD*DIVa and complex RHCE*CE-DIVa(2-3)-CE-D(5)-CE hybrid alleles (Figure 3), the latter expressing RH33, Riv, and FPTT (63).
On the basis of the data in the literature, it is thought that amino acids p.Leu169, p.Arg170 and p.Phe172 encoded by exon 4 in RHCE, and p.Glu233 encoded by exon 5 in RHD, are required on a single ‘hybrid protein’ for FPTT antigen expression. It is important to mention that amino acids 169, 170, and 172 reside within the extracellular vestibule (74,75), in which substitutions have been extensively shown to be associated with partial D phenotype (18,19).
Hybrid alleles and RH32 antigen expression
Expression of LFA RH32 was reported in cells presenting with the variant RN and partial DBT phenotypes (76-81). The molecular basis of RN involves the RHCE*Ce-D(4)-Ce hybrid allele at the RHCE locus, namely RHCE*02.10.01 (66) (Figure 3). Another RN allele, i.e., RHCE*02.10.02, on the same background, but including the templated p.Thr152Asn missense variant encoded by RHD exon 3 (RHCE*Ce-D(3:455-4)-Ce), was described in the same study, but has never been reported in the literature since. In addition to RH32, RN RBCs carry weak partial C and weak partial e antigens, and are positive for LFA DAK (RH54), but deficient for HFA Sec (RH46) (66,77,79). Two different DBT alleles altering the RHD locus have been described so far: RHD*DBT1 (*14.01) and *DBT2 (*14.02), respectively structured as RHD-CE(5-7)-D and RHD-CE(5-9)-D (64,65).
Interestingly, the nature of amino acids critical for RH32 expression is assumed to be the mirror of FPTT. Indeed, p.Met169, p.Met170 and p.Ile172 encoded by exon 4 in RHD, and p.Gln233 encoded by exon 5 in RHCE, are required on a single ‘hybrid protein’ for RH32 antigen expression.
DVI alleles and BARC antigen expression
In 1989, Lomas and Mougey reported a differential expression of the BARC antigen in RBCs presenting with a DVI phenotype (82). Indeed, RH:2,-3,4,5 (or Ccee) and RH:-2,3,4,5 (or ccEe) DVI RBCs typed BARC-positive and -negative, respectively, suggesting heterogeneity in the molecular bases guiding the DVI phenotype (80,82). In 1997, the molecular structure of the gene in DVI RBCs was re-examined by Avent and colleagues (83), who characterized two different hybrid alleles: RHD*DVI.1 (*06.01), structured as RHD-CE(4-5)-D, with RHCE exon 5 specific of the ‘E’ allele (i.e., encoding p.Pro226), was associated with BARC– DVI RBCs, as confirmed by independent studies (84,85); while RHD*DVI.2 (*06.02), structured as RHD-CE(4-6)-D, but with RHCE exon 5 specific of the ‘e’ allele (i.e., encoding p.Ala226), as previously published (10), was found in BARC+ DVI RBCs. Three other DVI alleles, all encoding p.Ala226, have been formally described thereafter (Figure 4). On BARC typing, RHD*DVI.3 (RHD-CE(3-6)-D) and *DVI.4 (RHD-CE(3-5)-D) were positive, while RHD*DVI.3.02 (RHD-CE(3-6)-D(A399T)) was not investigated (86-88).

In addition to the templated missense SNVs brought in the hybrid alleles by the RHCE-specific sequences, including p.Leu169, p.Arg170 and p.Phe172, these observations reinforced the statement that p.Ala226 is critical to BARC antigen expression (83).
Other hybrid alleles, LFA expression and RH antigen deficiency
Many microconversions and/or conversions of large genomic regions in either RHD or RHCE variant alleles have been associated with the expression of several other LFAs, but also deficiency in HFA. A nonexhaustive list of such examples is provided in Table 2.
Table 2
| Phenotype1 | Allele ID or designation | Molecular structure2 | References |
|---|---|---|---|
| RH:42 | RHD*DIIIa-CEVS(4-7)-D (*03N.01) | RHD(L62F,A137V,N152T)-CE(4-7)-D(G336C) | (89) |
| RH:54 | RHD*DIIIa (*03.01) | RHD(L62F,A137V,N152T,T201R,F223V) | (90-92) |
| RH:–18,–20 | RHCE*ceAR (*01.04.01) | RHCE(W16C,M238V,L245V,R263G,M267K,I306V) | (93,94) |
| RH:–18,49 | RHCE*ceBI (*01.08) | RHCE(W16C,M238V,A273V,L378V) | (15) |
| RH:43,–58 | RHCE*ceCF (*01.20.06) | RHCE(W16C,Q233E,L245V) | (95,96) |
| RH:–18,–34,–61 | RHCE*ceMO (*01.07.01) | RHCE(W16C,V223F) | (97,98) |
| RH:–18,49 | RHCE*ceSM (*01.09) | RHCE(W16C,M238V,A273V) | (99) |
| RH:20,–34 | RHCE*ceVS.01 (*01.20.01) | RHCE(L245V) | (100,101) |
| RH:56 | RHCE*CeNR (*02.08.02) | RHCE(Q41R)-D(6-10) | (102) |
| RH:–46,54 | RHCE*CeRN (*02.10.01) | RHCE-D(4)-CE | (66,91) |
| RH:33 | RHCE*CeVa (*02.04) | RHCE-D(5)-CE | (62) |
| RH:37 | N/A | RHCE-D(2-6)-CE | (14) |
1, phenotype regarding the expression of LFAs VS (RH20), RH33, Evans (RH37), RH42, Crawford (RH43), STEM (RH49), DAK (RH54), and CENR (RH56); and HFAs Hr (RH18), HrB (RH34), Sec (RH46), CELO (RH58), and RH61. 2, amino acid changes due to microconversions are underlined. LFA, low-frequency antigen; HFA, high-frequency antigen; N/A, not applicable.
Also, it is important to remind that, when gene conversion involves large genomic regions, including those encoding all epitopes, hybrid genes may result in the complete abolishment of D or CcEe antigen expression if occurring at the RHD or RHCE locus, respectively. As soon as in 1994, Blunt and colleagues reported the identification of RHD-specific sequences in D– donors of African origin (103). The authors hypothesized the presence of a recombinant RHD/CE gene in the so-called ‘dCes/dce’ genomes, which was later fully characterized and referred to as the (C)ces type 1 (*03N.01) allele (101,104). Several other D– hybrid alleles have been described since (Table 3).
Table 3
| Allele ID or designation | Molecular structure | References |
|---|---|---|
| RHD*03N.011 | RHD(L62F,A137V,N152T)-CE(4-7)-D(G336C) | (101,103,104) |
| RHD*01N.02 | RHCE(1-9)-D | (13) |
| RHD*01N.03 | RHD-CE(3-9)-D | (13,105) |
| RHD*01N.04 | RHD(S68T)-CE(3-9)-D | (17) |
| RHD*01N.05 | RHD-CE(3-7)-D | (13,17) |
| RHD*01N.062 | RHD-CE(4-7)-D(G336C) | (89) |
| RHD*01N.07 | RHD-CE(4-7)-D | (13,17,106) |
| RHD-CE(3-10)-D | RHD-CE(3-10) | (17,107) |
| RHD*01EL.443 | RHD-CE(4-9)-D | (108,109) |
Finally, hybrid genes in RHCE causing a complete deficiency in CcEe antigen expression have been less documented. However, beyond the alleles involved in the rare Dc– and DCW– phenotypes discussed above (11), investigation of rare D-- individuals has contributed to the identification of several variant alleles, including RHCE-D(3-7)-CE, RHCE-D(3-9)-CE, RHCE-D(4-9)-CE, RHD(1-9)-CE, and RHCE-D(2-6)-CE [reviewed in (110)], the latter expressing the Evans (RH37) antigen (14). Identification and registration of the donors and patients deficient in CcEe antigens are definitely critical for the management of these precious resources for future transfusions.
Molecular epidemiology of RH hybrid genes: a population-specific perspective with variable outcomes
Although RH hybrid genes have been found and characterized in all populations, allele structure and frequency are typically population-dependent with high specificity. Numerous molecular epidemiology studies have contributed to document and enrich the catalogue of RH molecular genetic polymorphism.
The African situation
In 1997, Daniels et al. reported that, while the RHD gene was systematically absent in serologically D– White Europeans, RHD-specific sequences were found in as many as 22/25 D– Black South Africans blood donors (111). For the first time, it was shown that the molecular basis of the D– phenotype is highly dependent on the ethnicity of the population of interest.
As indicated above, variant alleles from the DIVa and weak D type 4 clusters found in populations of African ancestry are basically hybrid alleles, including the D-negative (C)ces type 1 allele in the former cluster. The nonfunctional RHD pseudogene in the latter cluster, known as RHDψ (*08N.01) (112), also completes the definition. However, the D– phenotype induced by this allele is not directly due to the c.667T>G microconversion, but to a 37 bp-microduplication at intron 3/exon 4 junction and a putative premature stop codon, thus exhibiting a different mechanism resulting in a similar D– phenotype. Therefore, considering RHDψ as a ‘D– hybrid allele’ may be abusive in the strict sense. Importantly, it was initially described at various frequencies in serologically D– individuals of African ancestry: 54/82 (65.9%) in Black Africans from South Africa, Zimbabwe, and Ghana; 13/54 (24.1%) in African Americans; and 7/41 (17.1%) in mixed-race South Africans (112). In a preliminary study of 58 randomly selected blood donors from Mali, Wagner and colleagues identified (C)ces type 1 and RHDψ in five and seven samples, respectively, by exon-specific PCR using sequence-specific primers (PCR-SSP) and PCR-restriction fragment length polymorphism (PCR-RFLP), confirming that those alleles are commonly found in the general population in Africa (113). By using different techniques, other reports have shown that hybrid alleles are found in ~10-15% of the serologically D– donors of African origin (Table 4), although the data currently available have mostly involved a limited number of samples.
Table 4
| Country | Total number | RHD alleles (occurrence, %) | References | |||
|---|---|---|---|---|---|---|
| RHD*01N.011 | RHD*08N.012 | Hybrids3 | Others4 | |||
| South Africa/Ethiopia/Curaçao | 112 | 78 (69.6) | 15 (13.4) | 17 (15.2) | 2 (1.8) | (114) |
| Congo | 110 | 59 (53.6) | 35 (31.8) | 16 (14.6) | 0 | (46) |
| France (African origin5) | 36 | 24 (66.7) | 7 (19.4) | 4 (11.1) | 1 (2.8) | (115) |
See the respective references for information about sample selection. 1, *01N.01/*01N.01 (*01N.01 homozygous). 2, *08N.01/*08N.01 or *08N.01/*01N.01 (*08N.01 = RHDψ). 3, Including alleles from the DIVa (e.g., (C)ces type 1) and/or weak D type 4 clusters in trans with *08N.01. 4, Including SNVs. 5, Selected by their Fy(a–b–) phenotype.
But more importantly and as illustrated in the previous section, hybrid alleles are not confined to D-negativity, but have also been associated to a broad range of partial phenotypes. Molecular studies in D+ individuals from sub-Saharan areas have described the remarkable complexity and heterogeneity of both RH genes in Africans, including the prevalence of hybrid alleles. By allele-specific primer-PCR (ASP-PCR) and sequencing approaches, Touinssi and colleagues identified several partial RHD alleles involving gene conversions from the DIVa, DAU and weak D type 4 clusters in 8/40 D+ samples from the Teke ethnic group in Central Congo (46). Granier and collaborators reported comprehensively the variability of both the RHD and RHCE genes in a total of 347 pygmoid and nonpygmoid individuals from sub-Saharan populations, including Western, Central, and Eastern Africa (116). At the RHD locus, RHD*weak D type 4.2.0 (*DAR1.00) and *DIIIa (*03.01) were found to be the most common partial D-CE-D hybrid alleles in 41 (11.8%) and 23 (6.6%) samples, respectively. In RHCE, the total number of alleles that may be considered as ‘hybrid’ was very high, most of them involving microconversions. Beyond the ‘typical’ African RHCE*ce(48C) (*01.01), RHCE*ce(733G) (*01.20.01), and RHCE*ce(48C,733G) (*01.20.02) alleles found in a total of 230 samples (66.3%), the most prevalent hybrid CE allele was RHCE*ceAR (*01.04.01) observed in as many as 57 individuals (16.4%), followed by RHCE*ceTI (*01.02.01; 5.2%), RHCE*ceMO (*01.07.01; 4.0%), and RHCE*ces (*01.20.03; 3.7%) (Figure 5). Logically, those four later alleles, which encode partial antigens, have been frequently linked to RHD*DAR, *DIVa, *DAU0, as well as *DIIIa and (C)ces, respectively (93,94,98,104,116,117). Another study in 46 Fulani individuals from Mali carried out by the same group pointed out the high prevalence of the partial RN allele (allele frequency: 0.195) (118), which is known to be clinically relevant. Finally, it is worth mentioning that a hybrid RHCE*ce-D(9)-ce allele has been found to be cis-associated with the D-negative RHDψ allele systematically (119,120). It is still unknown whether or not this allele is clinically relevant.

These studies in various regions of sub-Saharan Africa have been of the greatest interest, but have been definitely too rare. It will be critical to investigate systematically more countries and populations in a near future that will undoubtedly result in very valuable findings. So far, the heterogeneous RH gene variability in the African population has been mostly addressed in large-scale studies performed ‘outside’ Africa. For example, in 806 individuals presenting with an altered RhCE antigen expression or producing anti-RhCE in France, ~80% being of African ancestry, the most common variant genotypes were RN/RN in 71 samples (8.8%), followed by ceMO/ceMO (n=15), (C)ces type 1/ceAR (14), RN/ceMO (13), (C)ces type 1/(C)ces type 1 (10), and ceAR/ceAR (7), as well as by many other alleles and combinations with a lower frequency (121). Several other studies have documented the complexity in RHD and/or RHCE, as well as the prevalence of hybrid genes, in populations of African ancestry in the USA, France, or Brazil, including in patients with sickle cell disease (SCD) (115,122-131), a clinically-relevant category of patients who are particularly prone to RH alloimmunization.
Beside the population studies, another line of evidence in favour of the higher frequency of templated SNVs, which are markers for hybrid alleles, in the African population, comes from both reference human genomic and blood group gene databases, such as gnomAD (132), Erythrogene (133), Blood Antigens and Blood Group Database (134). Although the data are not finely organized at a high resolution as a function of ethnicity/geography, they strongly support the preferential association of some SNVs with African ancestry. For example, allele frequency of c.667T>G (rs1053356) in RHD and c.733C>G (rs1053361) in RHCE, which are both associated with several hybrid alleles, is 0.1062 and 0.2303 in the latter population, respectively, while only 0.0010 and 0.0024 in non-Finnish Europeans (gnomAD v3.1.1), thus highlighting the variable distribution of those changes as a function of populations.
As expected, the distribution of hybrid alleles in North Africa differs significantly from sub-Saharan Africa. In Tunisia, D-CE-D hybrid alleles, including the clinically-relevant (C)ces haplotype(s), have been reported in 0.8-2.5% of serologically D– donors (135-139), and in as much as 23.0% of the subpopulation expressing C and/or E (140). Molecular typing by various methods has identified RHD*weak D type 4.0 as the most common variant RHD allele by far. Individuals carrying this allele were found in 34/2000 (1.7%) Tunisian and 11/4458 (0.25%) Moroccan donors with variant D phenotype (137,141,142). RHD*DBT1 and *DOL1 were reported once each (134,140). Investigation of RHCE has been rare in this subset of population. However, the partial RHCE*ceTI type 2-like, *ceVS.01, *ceVS.05, *ceMO and *RN hybrid alleles were identified in Moroccans (142).
The Caucasian situation
As well known in the molecular immunohematology laboratories and early illustrated by the investigation of serologically D– donors (111), but also in (variant) D+ donors, variability in RH genes is much lower in Caucasians than in Africans.
The most common partial D phenotype in Caucasians is DVI. Its prevalence in European populations, including in the Netherlands, UK, USA, Australia, Germany and Austria (143-149), which was determined by serological studies using monoclonal antibodies before the molecular bases of DVI phenotype were defined, has been known to be 0.02–0.05%. The five hybrid alleles resulting in DVI were initially identified in Caucasians, as indicated above. Molecular studies have shown a various distribution of allele frequency in different European regions. For example, RHD*DVI.1 and *DVI.2 are more prevalent in Germany, Austria, and Belgium, although with interregional variations (86,87,149), while *DVI.4 is the most common in Spain (88). Definitely many other clinically-relevant hybrid alleles at both the RHD and RHCE loci have been identified, characterized and reported in Caucasians, but to a much lower extent than DVI.
In 1997, Avent and collaborators investigated the molecular basis of serologically D–individuals from UK (150), which typically account for ~15% of the whole population in Europeans (151). By using a simple multiplex PCR approach targeting the RHD gene in this subgroup of individuals, they showed that 7 and 11 out of the 85 samples typed C+ and/or E+ (C/E+) were positive for intron 4 and exon 10 amplifications, respectively, whereas all 55 C–E– samples were negative. While the genotypes were not fully characterized at that time, the results clearly suggested that (I) D– phenotype is not strictly due to the homozygous deletion of the RHD gene in Caucasians and, interestingly, (II) nonfunctional hybrid alleles at the RHD locus are preferentially carried by individuals expressing C and/or E antigen(s). The later statement was subsequently confirmed in large-scale studies carried out in Central Europe. Indeed, D-CE-D hybrid allele frequency in serologically D– C/E+ Caucasians, including various large genomic conversions (Table 3), was estimated to be 0.006-0.018 (13,17,152).
The Asian situation
In East Asia, D+ phenotype is dramatically prevalent, typically >99% (151). In the remaining serologically D– individuals, Okuda and colleagues demonstrated that specific regions of the RHD gene were amplified by PCR in ~30% of Japanese donors, suggesting the presence of RHD variant alleles (153). Further investigations of such subsets in South Korea, China, Taiwan, and Japan, have identified hybrid alleles with gross rearrangements in ~8% (range, 2.8–16.6%), with a strong positive bias towards C/E+ individuals (153-165). RHD*01N.03 is usually the most common hybrid allele resulting in the so-called ‘true’ D– phenotype except in Japan, where RHD*01N.04 is more prevalent (Table 3). In the course of comprehensive large-scale and case studies, many hybrid alleles exhibiting a partial D phenotype have been reported, including RHD*DIV, *DV, *DVI, *DFR, *DBS, *DCS, *DBT, and *DLX (50,51,65,160,161,164,166-174). Among these, alleles from the DV and DVI (mainly RHD*06.03) categories appear to be the most common, although at a very low frequency, typically <10-5 (160,161,166,172,174).
Beside the numerous studies carried out in East Asia, comprehensive investigations in other Asian countries have been limited so far. In Thailand (Southeast Asia), where individuals presenting with a ‘non-D+’ phenotype are also <1% (D–: 0.30%; weak D: 0.01%) (175), a distribution comparable to that observed in East Asia has been found. In brief, ~11% of the serologically D– samples carry a hybrid gene, RHD*01N.03 being the most common; while RHD*06.03 is harboured by 24% of the weak/partial D donors in the Bangkok area, and even up to 37% at the nation-wide level (176,177). Finally, in India, where 3–7% of the whole population types D–, RHD*01N.03 is more frequent in C/E+ donors (29/171, 17.0%), followed by *01N.05 (8.8%) and *01EL.44 (2.9%) (109). Hybrid alleles appear to be rare and private in weak/partial D samples (178).
Analysis of the RHCE locus has not been carried out at a large-scale level so far, notably as variability in antigens carried by the RhCE protein and the related risk of alloimmunization is globally limited in Asia. Nonetheless some hybrid alleles have been reported, such as RHCE*D(1-3)-cE (*03.02) and RHCE*cE-D(5:697-712)-cE (*03.03), which both encode a partial E antigen (179,180), as well as other negative RHCE alleles identified in rare D-- individuals (110).
RH hybrid genes in daily practice: a clinical perspective
The key factors for an efficient molecular typing strategy
Transfusion medicine has been known as the pioneering discipline applying a personalized approach, which is logically based on both donor and patient ‘features’, herein blood group antigens carried by RBCs, and how they ‘match’ together. Therefore, efficient typing strategies are critical to this achievement. In immunohematology, the gold-standard for typing has remained serological testing. However, low- to medium-, and even high-throughput molecular approaches have been extensively implemented in laboratories as valuable complementary tools to gain into accurate prediction of the phenotype and optimization of patient safety, which is the ultimate goal (181-184). As illustrated above, hybrids and microconversions in RH genes are multiple, polymorphic, population-specific and confer a broad range of RH phenotypes with variable clinical interests. Therefore, the extent to which molecular testing is required as a complementary approach, as well as the strategy to be used thus basically depends on the status of the individual (donor or patient), his/her phenotype, ethnicity (African vs. Caucasian vs. Asian vs. admixed), as well as pathological status potentially.
Research and diagnostics in transfusion medicine
Since the identification of the RH genes, various approaches taking advantage of the successive technological advances in the fields of genetics and genomics have been designed and experienced. The simple PCR-SSP strategy, declined in both simplex and multiplex formats, followed by an agarose gel electrophoresis analysis, has been recognized for years as a simple and convenient approach to screen for the presence of specific RH segments in both transfusion and obstetrics, but also to genotype several hybrid alleles (86,185-195). As an example, identification of those clinically-relevant hybrid alleles in DVI individuals, who were proposed to be considered as ‘D-positive donor, D-negative recipient’ in as early as 1995 (26), was thus rapidly implemented in several specialized laboratories. Since the nineties, many low- to medium-throughput analytical methods, as well as commercial approaches, including genotyping platforms using PCR-based assays coupled with various detection systems for data acquisition, have been developed and shown to be robust to this purpose (183,196).
A significant advance in hybrid identification for the past ten years has been the implementation of strategies dedicated to the quantification of exon copy number variations (CNVs), namely the (commercial) multiplex ligation-dependent probe amplification (MLPA) (119) and the (in-house) quantitative multiplex PCR of short fluorescent fragments (QMPSF) (120), currently used in several laboratories (personal communications). Indeed, both cost-effective approaches have proven valuable and potent for the characterization of complex genotypes and alleles, notably in compound heterozygous conditions that had remained unresolved by conventional methods, such as PCR-SSP, Sanger sequencing and commercial microarrays (109,142,164,176-178).
Finally, for the very past years, the strategies that have shown great promise for blood group genotyping have been high-density DNA arrays and next-generation sequencing (NGS). Although the former has not formally proven yet its efficiency for resolving complex cases in RH to my knowledge, the latter has been used to investigate RHD zygosity initially, but also and more importantly for the identification of those hybrids in the RH genes. Indeed, since the preliminary NGS studies in the blood group field, which aimed to identify SNVs primarily (197-198), many investigators have addressed successfully the CNVs by using different platforms, as well as various strategies to generate [e.g., whole-genome sequencing (WGS), whole-exome sequencing (WES), and targeted-exome sequencing (TES)] and process the data (read depth analysis, automated software…) (128,131,134,199-205).
The challenge of patients with sickle cell disease (SCD)
Sickle cell disease (SCD, OMIM #603903) is a genetic blood disorder caused by variations in HBB, the gene encoding the β-globin polypeptide, a subunit of the oligomeric haemoglobin protein (206). RBC transfusion has been a common therapeutic strategy for the management of acute and chronic symptoms of the disease (207). Preventing alloimmunization in these multitransfused patients is thus a major critical challenge to guarantee their safety in future transfusions.
Patients with SCD are of African descent while, in Western countries, blood donors are mostly of European ancestry. This discrepancy in the respective origin of donors and patients illustrated by the differential qualitative and quantitative distributions of antigens between the individuals is critical (208). It has been well documented that patients with SCD, who are thus prone to be exposed regularly to foreign (donor) antigens, are far more at risk to alloimmunization following transfusion, but also to delayed hemolytic transfusion reaction (DHTR), than any other patients (209). In the RH system, antibodies involved in DHTR are frequently directed against C and E (~40%), which are less common in Africans (210).
Although there is no international consensus among the transfusion community, the strategy for transfusing patients with SCD that initially relied on the delivery of ABO/D-matched RBC units determined by serological testing progressively evolved towards extended matching for C, E, and K antigens (the latter from the Kell blood group system), which is standard practice currently, and for other antigens from the MNS (S), Duffy (Fya, Fyb), and Kidd (Jka, Jkb) blood group systems when possible. This trend towards extended RBC antigen-matching has helped to decrease dramatically the rate of alloimmunization in patients with SCD: from 18–75% with ABO/D only to 5–24% when extended to C/E/K, and up to 0–7% for optimal matching with additional antigens (211-213). Introduction of DNA-based approach for blood group genotyping has further contributed to facilitate transfusion support with extended antigen-matched blood, notably by predicting those RBC antigens that cannot be investigated by serological testing (214-219).
Even though RH antigen-matching has been carried out for the most common antigens, Chou and collaborators showed that production of RH alloantibodies occurred in 80/182 patients with SCD transfused primarily with RBC units from African American donors (123). Among the 146 antibodies found in the cohort, unexplained RH specificity was demonstrated for 91, including against D, C, E, and e, but also the Goa, V/VS, RH32, and CW LFAs, which are known to be due to RH variant alleles (see above). Upon molecular investigation of 226 patients with SCD in the RH genes, the authors showed a great allele diversity in both genes: 217 (48.0%) and 259 (57.3%) were found to be RHD and RHCE variant alleles, respectively, of which the majority involves hybrids alleles or microconversions, as previously reported in individuals of African descent (15,121,122,220,221). Importantly from a clinical point of view, those variant alleles are mostly associated with a partial antigen and/or LFA expression and/or HFA deficiency. Overall, this study elegantly confirmed the challenge of supplying RH antigen-matched RBC units to patients with SCD, even with donors originating from the same community. It also suggested to carry out RH genotyping in donors, additionally to patients, to improve identification of antigen-negative RBC units, thus increasing compatibility by molecular matching and reducing the risk of alloimmunization ultimately. The conclusions of this study were later supported by others (126,128,130). The latter strategy was further investigated in different laboratories, notably in the USA and Brazil, where the supply of compatible RBCs to patients with SCD is particularly challenging. All authors concluded invariably on the relevance and superior benefit of the prophylactic RH molecular matching over serological matching to optimize resources and delivery of compatible RBC units to patients with SCD, and recommended to promote blood donation in individuals of African descents (125,127,129,222-227). Implementation of this strategy routinely at a large-scale level, which requires high-throughput genotyping tools accompanied by bioinformatics resources and medical expertise, still remains a major challenge to achieve.
Conclusions
Since its discovery in the 1940s, the RH blood group system has been the matter of thousands of studies guided by the curiosity of clinicians, immunohematologists, biochemists, and latter geneticists and molecular biologists, as a fascinating example of transdisciplinary efforts towards its knowledge. RH molecular genetics is a complex field. Hybrid alleles and microconversions, not only from a genetic point of view, but also as the sources of the RH phenotypic variability through the expression of variable antigens and their clinical consequences, perfectly illustrate that complexity. Since the discovery of the RH genes 30 years ago, the successive advances in genetics and genomics, which have had a major impact in research and/or diagnostics, have contributed considerably to document the molecular repertoire at the origin of the variant phenotypes (184,228). Although most of the RH variant alleles can be resolved by the current tools nowadays, there is no doubt that other methods, including the third-generation (or long-read) sequencing technology that is becoming more and more available and attractive, will be key actors of future discoveries. At the population level, while numerous studies have been carried out, there is likely still much to learn in terms of RH molecular epidemiology in Africa, Asia, and many more isolated populations, but also many specific variant alleles to discover. Overall, while the current number of hybrid genes and microconversions has considerably increased since the characterization of the molecular basis of DVI category phenotype in 1994 (10), the catalogue is not completed yet.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Lilian Castilho) for the series “Serology and Molecular Biology of the Rh System” published in Annals of Blood. The article has undergone external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://aob.amegroups.com/article/view/10.21037/aob-21-55/coif). The series “Serology and Molecular Biology of the Rh System” was commissioned by the editorial office without any funding or sponsorship. YF serves as an unpaid editorial board member of Annals of Blood from July 2020 to July 2022. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the manuscript and ensure that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Avent ND, Ridgwell K, Tanner MJ, et al. cDNA cloning of a 30 kDa erythrocyte membrane protein associated with Rh (Rhesus)-blood-group-antigen expression. Biochem J 1990;271:821-5. [Crossref] [PubMed]
- Chérif-Zahar B, Bloy C, Le Van Kim C, et al. Molecular cloning and protein structure of a human blood group Rh polypeptide. Proc Natl Acad Sci U S A 1990;87:6243-7. [Crossref] [PubMed]
- Chérif-Zahar B, Mattéi MG, Le Van Kim C, et al. Localization of the human Rh blood group gene structure to chromosome region 1p34.3-1p36.1 by in situ hybridization. Hum Genet 1991;86:398-400. [Crossref] [PubMed]
- Colin Y, Chérif-Zahar B, Le Van Kim C, et al. Genetic basis of the RhD-positive and RhD-negative blood group polymorphism as determined by Southern analysis. Blood 1991;78:2747-52.
- Le van Kim C, Mouro I, Chérif-Zahar B, et al. Molecular cloning and primary structure of the human blood group RhD polypeptide. Proc Natl Acad Sci U S A 1992;89:10925-9. [Crossref] [PubMed]
- Arce MA, Thompson ES, Wagner S, et al. Molecular cloning of RhD cDNA derived from a gene present in RhD-positive, but not RhD-negative individuals. Blood 1993;82:651-5.
- Wagner FF, Flegel WA. RHCE represents the ancestral RH position, while RHD is the duplicated gene. Blood 2002;99:2272-3. [Crossref] [PubMed]
- Hermand P, Mouro I, Huet M, et al. Immunochemical characterization of rhesus proteins with antibodies raised against synthetic peptides. Blood 1993;82:669-76.
- Mouro I, Colin Y, Chérif-Zahar B, et al. Molecular genetic basis of the human Rhesus blood group system. Nat Genet 1993;5:62-5. [Crossref] [PubMed]
- Mouro I, Le Van Kim C, Rouillac C, et al. Rearrangements of the blood group RhD gene associated with the DVI category phenotype. Blood 1994;83:1129-35.
- Chérif-Zahar B, Raynal V, D'Ambrosio AM, et al. Molecular analysis of the structure and expression of the RH locus in individuals with D--, Dc-, and DCw- gene complexes. Blood 1994;84:4354-60.
- Wagner FF, Flegel WA. RHD gene deletion occurred in the Rhesus box. Blood 2000;95:3662-8.
- Wagner FF, Frohmajer A, Flegel WA. RHD positive haplotypes in D negative Europeans. BMC Genet 2001;2:10. [Crossref] [PubMed]
- Huang CH, Chen Y, Reid M, et al. Genetic recombination at the human RH locus: a family study of the red-cell Evans phenotype reveals a transfer of exons 2-6 from the RHD to the RHCE gene. Am J Hum Genet 1996;59:825-33.
- Noizat-Pirenne F, Lee K, Pennec PY, et al. Rare RHCE phenotypes in black individuals of Afro-Caribbean origin: identification and transfusion safety. Blood 2002;100:4223-31. [Crossref] [PubMed]
- Chen Q, Flegel WA. Random survey for RHD alleles among D+ European persons. Transfusion 2005;45:1183-91. [Crossref] [PubMed]
- Gassner C, Doescher A, Drnovsek TD, et al. Presence of RHD in serologically D-, C/E+ individuals: a European multicenter study. Transfusion 2005;45:527-38. [Crossref] [PubMed]
- Flegel WA, von Zabern I, Doescher A, et al. DCS-1, DCS-2, and DFV share amino acid substitutions at the extracellular RhD protein vestibule. Transfusion 2008;48:25-33. [Crossref] [PubMed]
- Flegel WA, von Zabern I, Doescher A, et al. D variants at the RhD vestibule in the weak D type 4 and Eurasian D clusters. Transfusion 2009;49:1059-69. [Crossref] [PubMed]
- von Zabern I, Wagner FF, Moulds JM, et al. D category IV: a group of clinically relevant and phylogenetically diverse partial D. Transfusion 2013;53:2960-73. [Crossref] [PubMed]
- Lomas C, Tippett P, Thompson KM, et al. Demonstration of seven epitopes on the Rh antigen D using human monoclonal anti-D antibodies and red cells from D categories. Vox Sang 1989;57:261-4. [Crossref] [PubMed]
- Tippett P. Serologically defined Rh determinants. J Immunogenet 1990;17:247-57. [Crossref] [PubMed]
- Tippett P, Moore S. Monoclonal antibodies against Rh and Rh related antigens. J Immunogenet 1990;17:309-19. [Crossref] [PubMed]
- Lomas C, McColl K, Tippett P. Further complexities of the Rh antigen D disclosed by testing category DII cells with monoclonal anti-D. Transfus Med 1993;3:67-9. [Crossref] [PubMed]
- Wagner FF, Frohmajer A, Ladewig B, et al. Weak D alleles express distinct phenotypes. Blood 2000;95:2699-708.
- Jones J, Scott ML, Voak D. Monoclonal anti-D specificity and Rh D structure: criteria for selection of monoclonal anti-D reagents for routine typing of patients and donors. Transfus Med 1995;5:171-84. [Crossref] [PubMed]
- Scott M. Section 1A: Rh serology. Coordinator's report. Transfus Clin Biol 2002;9:23-9. [Crossref] [PubMed]
- Floch A. Molecular genetics of the Rh blood group system: alleles and antibodies—a narrative review. Ann Blood 2021;6:29.
- Rouillac C, Colin Y, Hughes-Jones NC, et al. Transcript analysis of D category phenotypes predicts hybrid Rh D-CE-D proteins associated with alteration of D epitopes. Blood 1995;85:2937-44.
- Avent ND, Finning KM, Liu W, et al. Molecular biology of partial D phenotypes. Transfus Clin Biol 1996;3:511-6. [Crossref] [PubMed]
- Beckers EA, Faas BH, Ligthart P, et al. Characterization of the hybrid RHD gene leading to the partial D category IIIc phenotype. Transfusion 1996;36:567-74. [Crossref] [PubMed]
- Behzad O, Lee CL, Smith D. Hemolytic disease of the newborn due to anti-VS. Transfusion 1982;22:83. [Crossref] [PubMed]
- Spruell P, Lacey P, Bradford MF, et al. Incidence of hemolytic disease of the newborn due to anti-DW. Transfusion 1997;37:43S.
- Alter AA, Gelb AG, Lee SL. Hemolytic disease of the newborn caused by a new antibody (anti-Go(a)). Bibl Haematol 1964;19:341-3. [Crossref] [PubMed]
- Alter AA, Gelb AG, Chown B, et al. Gonzales (Goa), a new blood group character. Transfusion 1967;7:88-91. [Crossref] [PubMed]
- Leschek E, Pearlman SA, Boudreaux I, et al. Severe hemolytic disease of the newborn caused by anti-Gonzales antibody. Am J Perinatol 1993;10:362-4. [Crossref] [PubMed]
- Larson PJ, Lukas MB, Friedman DF, et al. Delayed hemolytic transfusion reaction due to anti-Go(a), an antibody against the low-prevalence Gonzales antigen. Am J Hematol 1996;53:248-50. [Crossref] [PubMed]
- Orlina AR, Unger PJ, Lacey PA. Anti-Rh32 causing severe hemolytic disease of the newborn. Rev Fr Transfus Immunohematol 1984;27:613-8. [Crossref] [PubMed]
- Issitt PD, Gutgsell NS, Martin PA, et al. Hemolytic disease of the newborn caused by anti-Rh32 and demonstration that RN encodes rhi (Ce,Rh7). Transfusion 1991;31:63-6. [Crossref] [PubMed]
- Toly-Ndour C, Huguet-Jacquot S, Pernot F, et al. Anticorps anti-privé dans le système RH et maladie hémolytique du nouveau-né: à propos de 2 cas. Transfus Clin Biol 2017;24:347.
- Contreras M, Stebbing B, Blessing M, et al. The Rh antigen Evans. Vox Sang 1978;34:208-11. [Crossref] [PubMed]
- Moulds JJ, Case J, Thornton S, et al. Anti-Ces: a previously undescribed Rh antibody. Transfusion 1980;20:631-2.
- Marais I, Moores P, Smart E, et al. STEM, a new low-frequency Rh antigen associated with the e-variant phenotypes hrS-(Rh: -18, -19) and hrB-(Rh: -31, -34). Transfus Med 1993;3:35-41. [Crossref] [PubMed]
- Lewis M, Chown B, Kaita H, et al. Blood group antigen Goa and the Rh system. Transfusion 1967;7:440-1. [Crossref] [PubMed]
- Chown B, Lewis M, Kaita H, et al. On the antigen Goa and the Rh system. Vox Sang 1968;15:264-71. [Crossref] [PubMed]
- Touinssi M, Chapel-Fernandes S, Granier T, et al. Molecular analysis of inactive and active RHD alleles in native Congolese cohorts. Transfusion 2009;49:1353-60. [Crossref] [PubMed]
- Wagner FF, Gassner C, Müller TH, et al. Molecular basis of weak D phenotypes. Blood 1999;93:385-93.
- Wagner FF, Gassner C, Eicher NI, et al. Characterization of D category IV type IV, DFW, and DNB. Transfusion 1998;38:63S.
- Hyodo H, Ishikawa Y, Tsuneyama H, et al. New RhD(IVb) identified in Japanese. Vox Sang 2000;79:116-7. [Crossref] [PubMed]
- Omi T, Takahashi J, Tsudo N, et al. The genomic organization of the partial D category DVa: the presence of a new partial D associated with the DVa phenotype. Biochem Biophys Res Commun 1999;254:786-94. [Crossref] [PubMed]
- Omi T, Okuda H, Iwamoto S, et al. Detection of Rh23 in the partial D phenotype associated with the D(Va) category. Transfusion 2000;40:256-8. [Crossref] [PubMed]
- Lopez GH, McGowan EC, McGrath KA, et al. A D+ blood donor with a novel RHD*D-CE(5-6)-D gene variant exhibits the low-frequency antigen RH23 (D(W)) characteristic of the partial DVa phenotype. Transfusion 2016;56:2322-30. [Crossref] [PubMed]
- Wagner FF, Ernst M, Sonneborn HH, et al. A D(V)-like phenotype is obliterated by A226P in the partial D DBS. Transfusion 2001;41:1052-8. [Crossref] [PubMed]
- Omi T, Takahashi J, Seno T, et al. Isolation, characterization, and family study of DTI, a novel partial D phenotype affecting the fourth external loop of D polypeptides. Transfusion 2002;42:481-9. [Crossref] [PubMed]
- Chown B, Lewis M, Kaita H. A new Rh antigen and antibody. Transfusion 1962;2:150-4. [Crossref] [PubMed]
- Bizot M, Lomas C, Rubio F, et al. An antiserum identifying a red cell determinant expressed by Rh:33 and by some “new” depressed Rh phenotypes. Transfusion 1988;28:342-5. [Crossref] [PubMed]
- omas C, Grässmann W, Ford D, et al. FPTT is a low-incidence Rh antigen associated with a “new” partial Rh D phenotype, DFR. Transfusion 1994;34:612-6.
- Faas BH, Beckers EA, Maaskant-van Wijk PA, et al. Molecular characterization of qualitative Rh variants. Biotest Bull 1997;5:439-49.
- von Zabern I, Flegel WA. IVS5-38del4 deletion in the RHD gene does not cause a DEL phenotype: relevance for RHD alleles including DFR-3. Transfusion 2007;47:1552-5. [Crossref] [PubMed]
- Ye L, Wang P, Gao H, et al. Partial D phenotypes and genotypes in the Chinese population. Transfusion 2012;52:241-6. [Crossref] [PubMed]
- Beckers EA, Faas BH, von dem Borne AE, et al. The R0Har RH:33 phenotype results from substitution of exon 5 of the RHCE gene by the corresponding exon of the RHD gene. Br J Haematol 1996;92:751-7. [Crossref] [PubMed]
- Noizat-Pirenne F, Le Pennec PY, Mouro I, et al. Molecular background of D(C)(e) haplotypes within the white population. Transfusion 2002;42:627-33. [Crossref] [PubMed]
- Hipsky CH, Hue-Roye K, Lomas-Francis C, et al. Molecular basis of the rare gene complex, DIVa(C)-, which encodes four low-prevalence antigens in the Rh blood group system. Vox Sang 2012;102:167-70. [Crossref] [PubMed]
- Beckers EA, Faas BH, Simsek S, et al. The genetic basis of a new partial D antigen: DDBT. Br J Haematol 1996;93:720-7. [Crossref] [PubMed]
- Huang CH, Chen Y, Reid ME, et al. Evidence for a separate genetic origin of the partial D phenotype DBT in a Japanese family. Transfusion 1999;39:1259-65. [Crossref] [PubMed]
- Rouillac C, Gane P, Cartron J, et al. Molecular basis of the altered antigenic expression of RhD in weak D(Du) and RhC/e in RN phenotypes. Blood 1996;87:4853-61.
- Giles CM, Crossland JD, Haggas WK, et al. An Rh gene complex which results in a "new" antigen detectable by a specific antibody, Anti-Rh 33. Vox Sang 1971;21:289-301. [Crossref] [PubMed]
- Beckers EA, Porcelijn L, Ligthart P, et al. The RoHar antigenic complex is associated with a limited number of D epitopes and alloanti-D production: a study of three unrelated persons and their families. Transfusion 1996;36:104-8. [Crossref] [PubMed]
- Wallace M, Lomas-Francis C, Tippett P. The D antigen characteristic of R0Har is a partial D antigen. Vox Sang 1996;70:169-72. [Crossref] [PubMed]
- Salmon C, Gerbal A, Liberge G, et al. The gene complex DIV (C)-. Rev Fr Transfus 1969;12:239-47. [Crossref] [PubMed]
- Habibi B, Perrier P, Salmon C. Antigen Nou. A new high frequency Rh antigen. Rev Fr Transfus Immunohematol 1981;24:117-20. [Crossref] [PubMed]
- Daniels GL. An investigation of the immune response of homozygotes for the Rh haplotype --D-- and related haplotypes. Using cells of rare Rh phenotypes. Rev Fr Transfus Immunohematol 1982;25:185-97. [Crossref] [PubMed]
- Delehanty CL, Wilkinson SL, Issitt PD, et al. A new low incidence Rh antigen. Transfusion 1983;23:410.
- Conroy MJ, Bullough PA, Merrick M, et al. Modelling the human rhesus proteins: implications for structure and function. Br J Haematol 2005;131:543-51. [Crossref] [PubMed]
- Callebaut I, Dulin F, Bertrand O, et al. Hydrophobic cluster analysis and modeling of the human Rh protein three-dimensional structures. Transfus Clin Biol 2006;13:70-84. [Crossref] [PubMed]
- Rosenfield RE, Haber GV, Schroeder R, et al. Problems in Rh typing as revealed by a single Negro family. Am J Hum Genet 1960;12:147-59.
- Chown B, Lewis M, Kaita H. The Rh system. An anomaly of inheritance, probably due to mutation. Vox Sang 1971;21:385-96.
- Chown B, Lewis M, Kaita H, et al. An unlinked modifier of Rh blood groups: effects when heterozygous and when homozygous. Am J Hum Genet 1972;24:623-37.
- Le Pennec PY, Rouger P, Klein MT, et al. A serologic study or red cells and sera from 18 Rh:32,-46 (RN/RN) persons. Transfusion 1989;29:798-802. [Crossref] [PubMed]
- Tippett P, Lomas-Francis C, Wallace M. The Rh antigen D: partial D antigens and associated low incidence antigens. Vox Sang 1996;70:123-31. [Crossref] [PubMed]
- Wallace M, Lomas-Francis C, Beckers E, et al. DBT: a partial D phenotype associated with the low-incidence antigen Rh32. Transfus Med 1997;7:233-8. [Crossref] [PubMed]
- Lomas C, Mougey R. Rh antigen D: variable expression in DVI phenotypes; a possible subdivision of category VI by a low frequency antigen. Transfusion 1989;29:14S.
- Avent ND, Liu W, Jones JW, et al. Molecular analysis of Rh transcripts and polypeptides from individuals expressing the DVI variant phenotype: an RHD gene deletion event does not generate All DVIccEe phenotypes. Blood 1997;89:1779-86.
- Huang CH. Human DVI category erythrocytes: correlation of the phenotype with a novel hybrid RhD-CE-D gene but not an internally deleted RhD gene. Blood 1997;89:1834-5.
- Maaskant-van Wijk PA, Beckers EA, van Rhenen DJ, et al. Evidence that the RHD(VI) deletion genotype does not exist. Blood 1997;90:1709-11.
- Wagner FF, Gassner C, Muller TH, et al. Three molecular structures cause rhesus D category VI phenotypes with distinct immunohematologic features. Blood 1998;91:2157-68.
- Van Sandt VS, Gassner C, Emonds MP, et al. RHD variants in Flanders, Belgium. Transfusion 2015;55:1411-7. [Crossref] [PubMed]
- Esteban R, Montero R, Flegel WA, et al. The D category VI type 4 allele is prevalent in the Spanish population. Transfusion 2006;46:616-23. [Crossref] [PubMed]
- Pham BN, Peyrard T, Juszczak G, et al. Heterogeneous molecular background of the weak C, VS+, hr B-, Hr B- phenotype in black persons. Transfusion 2009;49:495-504. [Crossref] [PubMed]
- Wagner FF, Ladewig B, Angert KS, et al. The DAU allele cluster of the RHD gene. Blood 2002;100:306-11. [Crossref] [PubMed]
- Reid ME, Storry JR, Sausais L, et al. DAK, a new low-incidence antigen in the Rh blood group system. Transfusion 2003;43:1394-7. [Crossref] [PubMed]
- Westhoff CM, Vege S, Halter-Hipsky C, et al. DIIIa and DIII Type 5 are encoded by the same allele and are associated with altered RHCE*ce alleles: clinical implications. Transfusion 2010;50:1303-11. [Crossref] [PubMed]
- Hemker MB, Ligthart PC, Berger L, et al. DAR, a new RhD variant involving exons 4, 5, and 7, often in linkage with ceAR, a new Rhce variant frequently found in African blacks. Blood 1999;94:4337-42.
- Peyrard T, Pham BN, Poupel S, et al. Alloanti-c/ce in a c+ceAR/Ce patient suggests that the rare RHCE ceAR allele (ceAR) encodes a partial c antigen. Transfusion 2009;49:2406-11. [Crossref] [PubMed]
- Flegel WA, Wagner FF, Chen Q, et al. The RHCE allele ceCF: the molecular basis of Crawford (RH43). Transfusion 2006;46:1334-42. [Crossref] [PubMed]
- Hipsky CH, Lomas-Francis C, Fuchisawa A, et al. RHCE*ceCF encodes partial c and partial e but not CELO, an antigen antithetical to Crawford. Transfusion 2011;51:25-31. [Crossref] [PubMed]
- Noizat-Pirenne F, Mouro I, Le Pennec PY, et al. Two new alleles of the RHCE gene in Black individuals: the RHce allele ceMO and the RHcE allele cEMI. Br J Haematol 2001;113:672-9. [Crossref] [PubMed]
- Westhoff CM, Vege S, Horn T, et al. RHCE*ceMO is frequently in cis to RHD*DAU0 and encodes a hr(S) -, hr(B) -, RH:-61 phenotype in black persons: clinical significance. Transfusion 2013;53:2983-9. [Crossref] [PubMed]
- Reid ME, Halter Hipsky C, Hue-Roye K, et al. The low-prevalence Rh antigen STEM (RH49) is encoded by two different RHCE*ce818T alleles that are often in cis to RHD*DOL. Transfusion 2013;53:539-44. [Crossref] [PubMed]
- Steers F, Wallace M, Johnson P, et al. Denaturing gradient gel electrophoresis: a novel method for determining Rh phenotype from genomic DNA. Br J Haematol 1996;94:417-21. [Crossref] [PubMed]
- Faas BH, Beckers EA, Wildoer P, et al. Molecular background of VS and weak C expression in blacks. Transfusion 1997;37:38-44. [Crossref] [PubMed]
- Westhoff CM, Storry JR, Walker P, et al. A new hybrid RHCE gene (CeNR) is responsible for expression of a novel antigen. Transfusion 2004;44:1047-51. [Crossref] [PubMed]
- Blunt T, Daniels G, Carritt B. Serotype switching in a partially deleted RHD gene. Vox Sang 1994;67:397-401. [Crossref] [PubMed]
- Daniels GL, Faas BH, Green CA, et al. The VS and V blood group polymorphisms in Africans: a serologic and molecular analysis. Transfusion 1998;38:951-8. [Crossref] [PubMed]
- Huang CH. Alteration of RH gene structure and expression in human dCCee and DCW-red blood cells: phenotypic homozygosity versus genotypic heterozygosity. Blood 1996;88:2326-33.
- Faas BH, Beckers EA, Simsek S, et al. Involvement of Ser103 of the Rh polypeptides in G epitope formation. Transfusion 1996;36:506-11. [Crossref] [PubMed]
- Shao CP, Xiong W. A new hybrid RHD-positive, D antigen-negative allele. Transfus Med 2004;14:185-6. [Crossref] [PubMed]
- Li Q, Hou L, Guo ZH, et al. Molecular basis of the RHD gene in blood donors with DEL phenotypes in Shanghai. Vox Sang 2009;97:139-46. [Crossref] [PubMed]
- Kulkarni SS, Gogri H, Parchure D, et al. RHD-Positive Alleles among D- C/E+ Individuals from India. Transfus Med Hemother 2018;45:173-7. [Crossref] [PubMed]
- Kulkarni S, Mishra G, Maru H, et al. Molecular characterization of rare D--/D-- variants in individuals of Indian origin. Blood Transfus 2022;20:59-65. [Crossref] [PubMed]
- Daniels G, Green C, Smart E. Differences between RhD-negative Africans and RhD-negative Europeans. Lancet 1997;350:862-3. [Crossref] [PubMed]
- Singleton BK, Green CA, Avent ND, et al. The presence of an RHD pseudogene containing a 37 base pair duplication and a nonsense mutation in africans with the Rh D-negative blood group phenotype. Blood 2000;95:12-8.
- Wagner FF, Moulds JM, Tounkara A, et al. RHD allele distribution in Africans of Mali. BMC Genet 2003;4:14. [Crossref] [PubMed]
- Grootkerk-Tax MG, Maaskant-van Wijk PA, van Drunen J, et al. he highly variable RH locus in nonwhite persons hampers RHD zygosity determination but yields more insight into RH-related evolutionary events. Transfusion 2005;45:327-37. [Crossref] [PubMed]
- Kappler-Gratias S, Auxerre C, Dubeaux I, et al. Systematic RH genotyping and variant identification in French donors of African origin. Blood Transfus 2014;12:s264-72. [Crossref] [PubMed]
- Granier T, Beley S, Chiaroni J, et al. A comprehensive survey of both RHD and RHCE allele frequencies in sub-Saharan Africa. Transfusion 2013;53:3009-17. [Crossref] [PubMed]
- Westhoff CM, Vege S, Halter Hipsky C, et al. RHCE*ceTI encodes partial c and partial e and is often in cis to RHD*DIVa. Transfusion 2013;53:741-6. [Crossref] [PubMed]
- Ba A, Beley S, Chiaroni J, et al. RH diversity in Mali: characterization of a new haplotype RHD*DIVa/RHCE*ceTI(D2). Transfusion 2015;55:1423-31. [Crossref] [PubMed]
- Haer-Wigman L, Veldhuisen B, Jonkers R, et al. RHD and RHCE variant and zygosity genotyping via multiplex ligation-dependent probe amplification. Transfusion 2013;53:1559-74. [Crossref] [PubMed]
- Fichou Y, Le Maréchal C, Bryckaert L, et al. A convenient qualitative and quantitative method to investigate RHD-RHCE hybrid genes. Transfusion 2013;53:2974-82. [Crossref] [PubMed]
- Pham BN, Peyrard T, Juszczak G, et al. Analysis of RhCE variants among 806 individuals in France: considerations for transfusion safety, with emphasis on patients with sickle cell disease. Transfusion 2011;51:1249-60. [Crossref] [PubMed]
- Silvy M, Di Cristofaro J, Beley S, et al. Identification of RHCE and KEL alleles in large cohorts of Afro-Caribbean and Comorian donors by multiplex SNaPshot and fragment assays: a transfusion support for sickle cell disease patients. Br J Haematol 2011;154:260-70. [Crossref] [PubMed]
- Chou ST, Jackson T, Vege S, et al. High prevalence of red blood cell alloimmunization in sickle cell disease despite transfusion from Rh-matched minority donors. Blood 2013;122:1062-71. [Crossref] [PubMed]
- Arnoni CP, Latini FR, Muniz JG, et al. How do we identify RHD variants using a practical molecular approach? Transfusion 2014;54:962-9. [Crossref] [PubMed]
- Reid ME, Halter Hipsky C, Hue-Roye K, et al. Genomic analyses of RH alleles to improve transfusion therapy in patients with sickle cell disease. Blood Cells Mol Dis 2014;52:195-202. [Crossref] [PubMed]
- Sippert E, Fujita CR, Machado D, et al. Variant RH alleles and Rh immunisation in patients with sickle cell disease. Blood Transfus 2015;13:72-7. [Crossref] [PubMed]
- Gaspardi AC, Sippert EA, De Macedo MD, et al. Clinically relevant RHD-CE genotypes in patients with sickle cell disease and in African Brazilian donors. Blood Transfus 2016;14:449-54. [Crossref] [PubMed]
- Dezan MR, Ribeiro IH, Oliveira VB, et al. RHD and RHCE genotyping by next-generation sequencing is an effective strategy to identify molecular variants within sickle cell disease patients. Blood Cells Mol Dis 2017;65:8-15. [Crossref] [PubMed]
- Chou ST, Evans P, Vege S, et al. RH genotype matching for transfusion support in sickle cell disease. Blood 2018;132:1198-207. [Crossref] [PubMed]
- Dinardo CL, Kelly S, Dezan MR, et al. Diversity of RH and transfusion support in Brazilian sickle cell disease patients with unexplained Rh antibodies. Transfusion 2019;59:3228-35. [Crossref] [PubMed]
- Wheeler MM, Lannert KW, Huston H, et al. Genomic characterization of the RH locus detects complex and novel structural variation in multi-ethnic cohorts. Genet Med 2019;21:477-86. [Crossref] [PubMed]
- Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020;581:434-43. [Crossref] [PubMed]
- Möller M, Jöud M, Storry JR, et al. Erythrogene: a database for in-depth analysis of the extensive variation in 36 blood group systems in the 1000 Genomes Project. Blood Adv 2016;1:240-9. [Crossref] [PubMed]
- Lane WJ, Westhoff CM, Gleadall NS, et al. Automated typing of red blood cell and platelet antigens: a whole-genome sequencing study. Lancet Haematol 2018;5:e241-51. [Crossref] [PubMed]
- Kacem N, Chakroun T, Moussa H, et al. RHD zygosity assignments based on most probable genotype and hybrid Rhesus box detection in Tunisia. Transfus Med 2012;22:362-6. [Crossref] [PubMed]
- Moussa H, Tsochandaridis M, Chakroun T, et al. Molecular background of D-negative phenotype in the Tunisian population. Transfus Med 2012;22:192-8. [Crossref] [PubMed]
- Ouchari M, Jemni-Yaacoub S, Chakroun T, et al. RHD alleles in the Tunisian population. Asian J Transfus Sci 2013;7:119-24. [Crossref] [PubMed]
- Moussa H, Ghommen N, Romdhane H, et al. (C)ce(s) haplotype screening in Tunisian blood donors. Blood Transfus 2014;12:405-9. [Crossref] [PubMed]
- Sassi A, Ouchari M, Houissa B, et al. RHD genotyping and its implication in transfusion practice. Transfus Apher Sci 2014;51:59-63. [Crossref] [PubMed]
- Moussa H, Tsochandaridis M, Kacem N, et al. RHD positive among C/E+ and D-negative blood donors in Tunisia. Transfus Clin Biol 2014;21:320-3. [Crossref] [PubMed]
- Ouchari M, Romdhane H, Chakroun T, et al. Weak D in the Tunisian population. Blood Transfus 2015;13:295-301. [Crossref] [PubMed]
- El Housse H, El Wafi M, Ouabdelmoumene Z, et al. Comprehensive phenotypic and molecular investigation of RhD and RhCE variants in Moroccan blood donors. Blood Transfus 2019;17:151-6. [Crossref] [PubMed]
- van Rhenen DJ, Overbeeke MA. Quality control of anti-D sera by a panel of donor red cells with weak reacting D antigen and with partial D antigens by the Federation of Netherlands Red Cross Blood Banks. Vox Sang 1989;57:273-4. [Crossref] [PubMed]
- Leader KA, Kumpel BM, Poole GD, et al. Human monoclonal anti-D with reactivity against category DVI cells used in blood grouping and determination of the incidence of the category DVI phenotype in the DU population. Vox Sang 1990;58:106-11. [Crossref] [PubMed]
- Beck ML, Hardman JT. Incidence of D category VI among DU donors in the USA. Transfusion 1991;31:25S.
- Watt J. The incidence of category VI amongst weak Rh(D) positive Sydney blood donors. Transfus Med 1993;3:72.
- van Rhenen DJ, Thijssen PM, Overbeeke MA. Serological characteristics of partial D antigen category VI in 8 unrelated blood donors. Vox Sang 1994;66:133-6. [Crossref] [PubMed]
- Wagner FF, Kasulke D, Kerowgan M, et al. Frequencies of the blood groups ABO, Rhesus, D category VI, Kell, and of clinically relevant high-frequency antigens in south-western Germany. Infusionsther Transfusionsmed 1995;22:285-90. [Crossref] [PubMed]
- Polin H, Danzer M, Hofer K, et al. Effective molecular RHD typing strategy for blood donations. Transfusion 2007;47:1350-5. [Crossref] [PubMed]
- Avent ND, Martin PG, Armstrong-Fisher SS, et al. Evidence of genetic diversity underlying Rh D-, weak D (Du), and partial D phenotypes as determined by multiplex polymerase chain reaction analysis of the RHD gene. Blood 1997;89:2568-77.
- Daniels G. Rh and RHAG blood group systems. In: Daniels G. editor. Human Blood Groups. 3rd ed. Oxford: Blackwell, 2013:182-258.
- Gowland P, Gassner C, Hustinx H, et al. Molecular RHD screening of RhD negative donors can replace standard serological testing for RhD negative donors. Transfus Apher Sci 2014;50:163-8. [Crossref] [PubMed]
- Okuda H, Kawano M, Iwamoto S, et al. The RHD gene is highly detectable in RhD-negative Japanese donors. J Clin Invest 1997;100:373-9. [Crossref] [PubMed]
- Sun CF, Chou CS, Lai NC, et al. RHD gene polymorphisms among RhD-negative Chinese in Taiwan. Vox Sang 1998;75:52-7.
- Shao CP, Maas JH, Su YQ, et al. Molecular background of Rh D-positive, D-negative, D(el) and weak D phenotypes in Chinese. Vox Sang 2002;83:156-61. [Crossref] [PubMed]
- Peng CT, Shih MC, Liu TC, et al. Molecular basis for the RhD negative phenotype in Chinese. Int J Mol Med 2003;11:515-21.
- Kim JY, Kim SY, Kim CA, et al. Molecular characterization of D- Korean persons: development of a diagnostic strategy. Transfusion 2005;45:345-52. [Crossref] [PubMed]
- Xu Q, Grootkerk-Tax MG, Maaskant-van Wijk PA, et al. Systemic analysis and zygosity determination of the RHD gene in a D-negative Chinese Han population reveals a novel D-negative RHD gene. Vox Sang 2005;88:35-40. [Crossref] [PubMed]
- Luettringhaus TA, Cho D, Ryang DW, et al. An easy RHD genotyping strategy for D- East Asian persons applied to Korean blood donors. Transfusion 2006;46:2128-37. [Crossref] [PubMed]
- Ye LY, Guo ZH, Li Q, et al. Molecular and family analyses revealed two novel RHD alleles in a survey of a Chinese RhD-negative population. Vox Sang 2007;92:242-6. [Crossref] [PubMed]
- Ye SH, Wu DZ, Wang MN, et al. A comprehensive investigation of RHD polymorphisms in the Chinese Han population in Xi’an. Blood Transfus 2014;12:396-404. [Crossref] [PubMed]
- Ogasawara K, Suzuki Y, Sasaki K, et al. Molecular basis for D- Japanese: identification of novel DEL and D- alleles. Vox Sang 2015;109:359-65. [Crossref] [PubMed]
- Seo MH, Won EJ, Hong YJ, et al. An effective diagnostic strategy for accurate detection of RhD variants including Asian DEL type in apparently RhD-negative blood donors in Korea. Vox Sang 2016;111:425-30. [Crossref] [PubMed]
- Ji YL, Luo H, Wen JZ, et al. RHD genotype and zygosity analysis in the Chinese Southern Han D+, D- and D variant donors using the multiplex ligation-dependent probe amplification assay. Vox Sang 2017;112:660-70. [Crossref] [PubMed]
- Kim B, Lee ST, Kim S, et al. Application of Multiplex Ligation-Dependent Probe Amplification Assay for Genotyping Major Blood Group Systems Including DEL Variants in the D-Negative Korean Population. Ann Lab Med 2018;38:32-8. [Crossref] [PubMed]
- Chung YN, Kim TY, Yu H, et al. Molecular basis of serological weak D phenotypes and RhD typing discrepancies identified in the Korean population. Blood Transfus 2021;19:327-34. [Crossref] [PubMed]
- Hyodo H, Ishikawa Y, Kashiwase K, et al. Polymorphisms of RhD(Va) and a new RhD(Va)-like variant found in Japanese individuals. Vox Sang 2000;78:122-5. [Crossref] [PubMed]
- Legler TJ, Wiemann V, Ohto H, et al. D(Va) category phenotype and genotype in Japanese families. Vox Sang 2000;78:194-7. [Crossref] [PubMed]
- Lee YL, Chiou HL, Hu SN, et al. Analysis of RHD genes in Taiwanese RhD-negative donors by the multiplex PCR method. J Clin Lab Anal 2003;17:80-4. [Crossref] [PubMed]
- Tanaka M, Kamada I, Takahashi J, et al. Evaluation of a blood group genotyping platform (BLOODchip(®) Reference) in Japanese samples. Transfus Med 2014;24:39-44. [Crossref] [PubMed]
- He J, Ying Y, Hong X, et al. Molecular basis and zygosity determination of D variants including identification of four novel alleles in Chinese individuals. Transfusion 2015;55:137-43. [Crossref] [PubMed]
- Zhang X, Li G, Zhou Z, et al. Molecular and computational analysis of 45 samples with a serologic weak D phenotype detected among 132,479 blood donors in northeast China. J Transl Med 2019;17:393. [Crossref] [PubMed]
- Choi S, Yu H, Cho D. First Korean Case of Partial D DBS-1. Ann Lab Med 2020;40:337-40. [Crossref] [PubMed]
- Jeong D, Oh S, Song EY, et al. Molecular Characteristics of the Serological Weak D Phenotype in Koreans. Diagnostics (Basel) 2021;11:920. [Crossref] [PubMed]
- Fongsarun J, Nuchprayoon I, Yod-in S, et al. Blood groups in Thai blood donors. Thai J Hematol Transfus Med 2002;12:277-86.
- Thongbut J, Raud L, Férec C, et al. Comprehensive Molecular Analysis of Serologically D-Negative and Weak/Partial D Phenotype in Thai Blood Donors. Transfus Med Hemother 2020;47:54-60. [Crossref] [PubMed]
- Thongbut J, Laengsri V, Raud L, et al. Nation-wide investigation of RHD variants in Thai blood donors: Impact for molecular diagnostics. Transfusion 2021;61:931-8. [Crossref] [PubMed]
- Fichou Y, Parchure D, Gogri H, et al. Molecular basis of weak D expression in the Indian population and report of a novel, predominant variant RHD allele. Transfusion 2018;58:1540-9. [Crossref] [PubMed]
- Noizat-Pirenne F, Mouro I, Gane P, et al. Heterogeneity of blood group RhE variants revealed by serological analysis and molecular alteration of the RHCE gene and transcript. Br J Haematol 1998;103:429-36. [Crossref] [PubMed]
- Kashiwase K, Ishikawa Y, Hyodo H, et al. E variants found in Japanese and c antigenicity alteration without substitution in the second extracellular loop. Transfusion 2001;41:1408-12. [Crossref] [PubMed]
- Flegel WA, von Zabern I, Wagner FF. Six years' experience performing RHD genotyping to confirm D- red blood cell units in Germany for preventing anti-D immunizations. Transfusion 2009;49:465-71. [Crossref] [PubMed]
- Peyrard T. Use of genomics for decision-making in transfusion medicine: laboratory practice. ISBT Sci Ser 2013;8:11-5.
- Peyrard T. Molecular tools for investigating immunohaematology problems. ISBT Sci Ser 2015;10:31-8.
- Westhoff CM. Blood group genotyping. Blood 2019;133:1814-20. [Crossref] [PubMed]
- Bennett PR, Le Van Kim C, Colin Y, et al. Prenatal determination of fetal RhD type by DNA amplification. N Engl J Med 1993;329:607-10. [Crossref] [PubMed]
- Lo YM, Bowell PJ, Selinger M, et al. Prenatal determination of fetal RhD status by analysis of peripheral blood of rhesus negative mothers. Lancet 1993;341:1147-8. [Crossref] [PubMed]
- Pope J, Navarrete C, Warwick R, et al. Multiplex PCR analysis of RhD gene. Lancet 1995;346:375-6. [Crossref] [PubMed]
- Simsek S, Bleeker PM, von dem Borne AE. Prenatal determination of fetal RhD type. N Engl J Med 1994;330:795-6. [Crossref] [PubMed]
- Simsek S, Faas BH, Bleeker PM, et al. Rapid RhD genotyping by polymerase chain reaction based amplification of DNA. Blood 1995;85:2975-80.
- Gassner C, Schmarda A, Kilga-Nogler S, et al. RHD/CE typing by polymerase chain reaction using sequence-specific primers. Transfusion 1997;37:1020-6. [Crossref] [PubMed]
- Flegel WA, Wagner FF, Müller TH, et al. Rh phenotype prediction by DNA typing and its application to practice. Transfus Med 1998;8:281-302. [Crossref] [PubMed]
- Legler TJ, Maas JH, Blaschke V, et al. RHD genotyping in weak D phenotypes by multiple polymerase chain reactions. Transfusion 1998;38:434-40. [Crossref] [PubMed]
- Maaskant-van Wijk PA, Faas BH, de Ruijter JA, et al. Genotyping of RHD by multiplex polymerase chain reaction analysis of six RHD-specific exons. Transfusion 1998;38:1015-21. [Crossref] [PubMed]
- Cotorruelo C, Biondi C, Borrás SG, et al. Molecular determination of RhD phenotype by DNA typing: clinical applications. Ann Clin Biochem 2000;37:781-9. [Crossref] [PubMed]
- Cotorruelo CM, Biondi CS, Borrás SE, et al. A Dc- phenotype encoded by an RHCE-D(5-7/8)-CE hybrid allele. Vox Sang 2003;85:102-8. [Crossref] [PubMed]
- Tournamille C. Molecular biology methods in immunohematology. Transfus Clin Biol 2013;20:72-9. [Crossref] [PubMed]
- Stabentheiner S, Danzer M, Niklas N, et al. Overcoming methodical limits of standard RHD genotyping by next-generation sequencing. Vox Sang 2011;100:381-8. [Crossref] [PubMed]
- Fichou Y, Audrézet MP, Guéguen P, et al. Next-generation sequencing is a credible strategy for blood group genotyping. Br J Haematol 2014;167:554-62. [Crossref] [PubMed]
- Baronas J, Westhoff CM, Vege S, et al. RHD zygosity determination from whole genome sequencing data. J Blood Disorder Transfus 2016;7:5.
- Chou ST, Flanagan JM, Vege S, et al. Whole-exome sequencing for RH genotyping and alloimmunization risk in children with sickle cell anemia. Blood Adv 2017;1:1414-22. [Crossref] [PubMed]
- Schoeman EM, Lopez GH, McGowan EC, et al. Evaluation of targeted exome sequencing for 28 protein-based blood group systems, including the homologous gene systems, for blood group genotyping. Transfusion 2017;57:1078-88. [Crossref] [PubMed]
- Lane WJ, Vege S, Mah HH, et al. Automated typing of red blood cell and platelet antigens from whole exome sequences. Transfusion 2019;59:3253-63. [Crossref] [PubMed]
- Chang TC, Haupfear KM, Yu J, et al. A novel algorithm comprehensively characterizes human RH genes using whole-genome sequencing data. Blood Adv 2020;4:4347-57. [Crossref] [PubMed]
- Halls JBL, Vege S, Simmons DP, et al. Overcoming the challenges of interpreting complex and uncommon RH alleles from whole genomes. Vox Sang 2020;115:790-801. [Crossref] [PubMed]
- Stef M, Fennell K, Apraiz I, et al. RH genotyping by nonspecific quantitative next-generation sequencing. Transfusion 2020;60:2691-701. [Crossref] [PubMed]
- Rees DC, Williams TN, Gladwin MT. Sickle-cell disease. Lancet 2010;376:2018-31. [Crossref] [PubMed]
- Yazdanbakhsh K, Ware RE, Noizat-Pirenne F. Red blood cell alloimmunization in sickle cell disease: pathophysiology, risk factors, and transfusion management. Blood 2012;120:528-37. [Crossref] [PubMed]
- Vichinsky EP, Earles A, Johnson RA, et al. Alloimmunization in sickle cell anemia and transfusion of racially unmatched blood. N Engl J Med 1990;322:1617-21. [Crossref] [PubMed]
- Garratty G. Severe reactions associated with transfusion of patients with sickle cell disease. Transfusion 1997;37:357-61. [Crossref] [PubMed]
- Daniels G. Variants of RhD – current testing and clinical consequences. Br J Haematol 2013;161:461-70. [Crossref] [PubMed]
- LaSalle-Williams M, Nuss R, Le T, et al. Extended red blood cell antigen matching for transfusions in sickle cell disease: a review of a 14-year experience from a single center. Transfusion 2011;51:1732-9. [Crossref] [PubMed]
- Chou ST, Liem RI, Thompson AA. Challenges of alloimmunization in patients with haemoglobinopathies. Br J Haematol 2012;159:394-404. [Crossref] [PubMed]
- Fasano RM, Chou ST. Red Blood Cell Antigen Genotyping for Sickle Cell Disease, Thalassemia, and Other Transfusion Complications. Transfus Med Rev 2016;30:197-201. [Crossref] [PubMed]
- Ribeiro KR, Guarnieri MH, da Costa DC, et al. DNA array analysis for red blood cell antigens facilitates the transfusion support with antigen-matched blood in patients with sickle cell disease. Vox Sang 2009;97:147-52. [Crossref] [PubMed]
- Wilkinson K, Harris S, Gaur P, et al. Molecular blood typing augments serologic testing and allows for enhanced matching of red blood cells for transfusion in patients with sickle cell disease. Transfusion 2012;52:381-8. [Crossref] [PubMed]
- Belsito A, Costa D, Fiorito C, et al. Erythrocyte genotyping for transfusion-dependent patients at the Azienda Universitaria Policlinico of Naples. Transfus Apher Sci 2015;52:72-7. [Crossref] [PubMed]
- Casas J, Friedman DF, Jackson T, et al. Changing practice: red blood cell typing by molecular methods for patients with sickle cell disease. Transfusion 2015;55:1388-93. [Crossref] [PubMed]
- Karafin MS, Field JJ, Gottschall JL, et al. Barriers to using molecularly typed minority red blood cell donors in support of chronically transfused adult patients with sickle cell disease. Transfusion 2015;55:1399-406. [Crossref] [PubMed]
- Chou ST, Westhoff CM. Application of genomics for transfusion therapy in sickle cell anemia. Blood Cells Mol Dis 2017;67:148-54. [Crossref] [PubMed]
- Castilho L, Rios M, Rodrigues A, et al. High frequency of partial DIIIa and DAR alleles found in sickle cell disease patients suggests increased risk of alloimmunization to RhD. Transfus Med 2005;15:49-55. [Crossref] [PubMed]
- Noizat-Pirenne F, Tournamille C. Relevance of RH variants in transfusion of sickle cell patients. Transfus Clin Biol 2011;18:527-35. [Crossref] [PubMed]
- da Costa DC, Pellegrino J Jr, Guelsin GA, et al. Molecular matching of red blood cells is superior to serological matching in sickle cell disease patients. Rev Bras Hematol Hemoter 2013;35:35-8. [Crossref] [PubMed]
- Castilho L, Dinardo CL. Optimized Antigen-Matched in Sickle Cell Disease Patients: Chances and Challenges in Molecular Times - the Brazilian Way. Transfus Med Hemother 2018;45:258-62. [Crossref] [PubMed]
- Cruz BR, de Souza Silva TC, de Souza Castro B, et al. Molecular matching for patients with haematological diseases expressing altered RHD-RHCE genotypes. Vox Sang 2019;114:605-15. [Crossref] [PubMed]
- Santos TD, Macedo MD, Menegati SFP, et al. Challenges in providing compatible blood with Rh genotype-matching in Brazilian patients with sickle cell disease. Transfus Med 2019;29:423-9. [Crossref] [PubMed]
- Van Buren NL, Gorlin JB, Corby SM, et al. How do I incorporate red cell genotyping to improve chronic transfusion therapy? Transfusion 2020;60:16-25. [Crossref] [PubMed]
- Muniz JG, Arnoni C, Medeiros R, et al. Antigen matching for transfusion support in Brazilian female patients with sickle cell disease to reduce RBC alloimmunization. Transfusion 2021;61:2458-67. [Crossref] [PubMed]
- Hyland CA, Roulis EV, Schoeman EM. Developments beyond blood group serology in the genomics era. Br J Haematol 2019;184:897-911. [Crossref] [PubMed]
Cite this article as: Fichou Y. Hybrids and microconversions in RH genes: investigation and implication in transfusion therapy. Ann Blood 2023;8:7.


