
Transfusion transmitted bacterial infections (TTBI) involving contaminated platelet concentrates: residual risk despite intervention strategies
Introduction
Transfusion transmitted bacterial infection (TTBI) due to contamination of platelets is a major risk of blood transfusion (1,2). Measures to decrease the degree of platelet contamination include skin disinfection, diversion of the first blood flow after skin puncture, bacterial screening of blood products and application of pathogen reduction technology. Diversion of the first flow, in combination with an optimized disinfection method, has been shown to reduce initially positive cultures of pooled platelet concentrates (PC) from 0.95% to 0.37% (3). This finding led to the introduction of the diversion pouch in the Netherlands in 2004 and is currently common practice in blood banks across the world (3-5). To detect remaining bacterial contamination, various bacterial testing approaches are used, including culture-based or rapid detection tests. In many countries, including the Netherlands, pooled PC and apheresis platelets are tested for bacterial contamination by culturing a sample in the BacT/ALERT® system (6). The effectiveness of this approach is dependent on multiple parameters, including number and type of bottles, sampling volume and timing of sampling (7). Recommended strategies by the Food and Drug Administration (FDA) guideline on the safety of platelets include large volume delayed sampling, for example, applied in the United Kingdom (8,9).
Whereas these intervention strategies have significantly decreased septic transfusion reactions from bacterial contamination, a residual risk remains. In the European Union, all serious transfusion reactions possibly related to the quality and safety of blood products are notifiable and reported annually to the European Commission. In our country, Transfusion and Transplantation Reactions in Patients (TRIP) registers reports of transfusion reactions of all severity levels (passive surveillance). The TRIP database provides the opportunity to analyse reported cases of post-transfusion bacteremia/sepsis that were judged to be TTBI. In this study, we examined all cases of TTBI associated with platelet transfusions in 2008–2019 and reviewed the corresponding bacterial screening results of the platelet products.
In addition, we used a second approach to detect the remaining risk of TTBI due to contaminated platelets. PC are released as “negative-to-date” during BacT/ALERT® incubation. Whereas growth of bacteria is usually detectable during the time period between sampling and transfusion, transfusion does occur of platelets which subsequently have a positive result during the 7-day BacT/ALERT® incubation. In this study, we retrospectively examined transfusion reactions in cases in which platelets had been transfused and subsequently tested positive in the bacterial screening, during 2013–2019. Additionally, we characterized the bacterial species found in our bacterial screening.
Methods
Data on reported transfusion reactions
In the Netherlands, transfusion reactions, including those which may be related to the quality and/or safety of blood products are registered by TRIP, the national hemo- and biovigilance office (https://www.tripnet.nl/en/). TRIP defines post-transfusion bacteremia/sepsis as follows: clinical symptoms of bacteremia/sepsis arising during, directly after or some time subsequent to a blood transfusion, with a relevant positive patient blood culture result; a causal link to a transfused blood component may or may not be confirmed, through a finding of the same bacterial species in the component or material from the donor (10). TTBI is defined as a case of post-transfusion bacteremia/sepsis in which blood cultures of the recipient are positive with the same bacterial species as the culture from the transfused component; all information supplied by the hospital including resistance spectrum and occasionally genotyping is included. A team of experts within TRIP evaluates all TTBI cases and since 2011 has formally adjudicated the TTBI imputability, or likelihood that the reported septic reaction was caused by transmission of bacteria in the blood component (10). For this study, data on all TTBIs in 2008–2019 were provided by TRIP (11).
In addition, we reviewed transfusion reactions reported to TRIP in 2008–2019, other than TTBI, where bacteria were cultured from the remnant of the transfused platelet concentrate by the hospital while bacterial screening by the blood establishment remained negative.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The IRB ethical approval and individual informed consent are waived due to the retrospective nature of the study.
Bacterial screening with BacT/ALERT®
Buffy coats (BC) are produced from overnight hold whole blood (14–20 hours at room temperature) and pooled from 5 whole blood donations to produce a pooled BC-derived, leukocyte-depleted PC. Platelets are currently stored in platelet additive solution, but during 2013–2019 platelets were in plasma, platelet additive solution/plasma or platelet additive solution only, changing over the years. Products are sampled within 2 hours after production (i.e., 17–25 hours after donation) and anaerobic and aerobic blood culture bottles are inoculated (7.5 mL each) in a laminar flow cabinet. The bottles are incubated in the BacT/ALERT® (BioMerieux) system for a maximum of 7 days. Gram staining and culture are performed on bottles with a positive BacT/ALERT® signal. Confirmed positive results are defined as a positive BacT/ALERT® signal confirmed by a positive culture from the BacT/ALERT® bottle (majority of cases), or a positive Gram staining but a negative culture. Apheresis PC, which account for only about 7% of transfused PC in the Netherlands, are sampled within 12 hours after collection and are cultured according to the same procedure.
All platelet products are released to hospitals on a “negative-to-date” basis, i.e., the culture bottles are negative for bacterial growth at the moment of distributing the product. If the product becomes positive after release, the product is recalled and destroyed. However, the product may have been transfused already.
Positive bacterial screening after transfusion and description of cases
All results of our BacT/ALERT® screening, including both pooled BC derived PC as well as apheresis PC, are maintained in a database for quality control including trend analysis. In case of a positive BacT/ALERT® result of an already distributed product, hospitals are always informed and the product is recalled. If the PC has already been transfused, hospitals are asked whether any transfusion reaction occurred and whether blood cultures have been performed. These data are registered in our database using TrackWise Enterprise Quality Management Software (EQMS). The blood establishment assesses whether each transfusion reaction was possibly related to a contaminated PC.
For this study, BacT/ALERT® data, including number of confirmed positive results and number of transfused products, were obtained from our database for a 7-year period [2013–2019]. In addition, clinical data were obtained from TrackWise EQMS, including the assessment whether the transfusion reactions were potentially related to contaminated PC.
Characterisation of bacterial species found in the bacterial screening
We used our database of the BacT/ALERT® screening to characterise the distribution of bacterial species for all confirmed positives identified in the BacT/ALERT® during 2013–2019. The database includes information on time to positivity of the blood culture bottles. We analysed the contribution of bacterial species that were isolated within 48 hours or after 48 hours incubation. Analysis was restricted to pooled BC-derived platelets as the main platelet product.
Statistical analysis
No statistical analyses were performed.
Results
Post-transfusion bacteremia/sepsis and TTBI
In 2008–2019, 633 cases of post-transfusion bacteremia/sepsis were reported in the Netherlands, corresponding to an average of approximately 50 reactions per year. Of the 633 post-transfusion bacteremia/sepsis cases, only 23 cases met criteria for TTBI. Sixteen/23 TTBI cases involved PC. Seven/twenty-three cases occurred with red blood cell transfusion and were not included in this study. The total number of distributed PC during this time period was 668,896. Thus, TTBI occurred in 16 cases out of 668,896 distributed PC, corresponding to 1 in every 42,000 PCs (or 24 per 1,000,000 or 0.002%).
Characterisation of TTBI cases
The 16 TTBI cases associated with platelets were characterised (Table 1). A severity score was assigned to all TTBI cases, based on the International Haemovigilance Network/International Society for Blood Transfusion grading system. The severity grade was 1 (non-severe) in two cases, grade 2 (severe) in 11 cases, grade 3 (life-threatening) in two cases and grade 4 (death) in one case. The cultured bacterial species in the TTBI cases were the following: coagulase-negative staphylococci (7 cases), beta-hemolytic streptococci (3 cases), S. aureus (2 cases), E. faecalis, E. coli, Salmonella group B and Acinetobacter ursingii. TTBI imputability was assigned by TRIP experts based on clinical and microbiological information. There were two definite TTBI cases, six probable cases and eight possible cases (12).
Table 1
| Case number | Year | Severity grade | TTBI imputability | Symptoms | Patient blood culture results | Culture result from remnant of transfused blood component identical to patient blood culture† |
|---|---|---|---|---|---|---|
| 1 | 2008 | 2 | Possible | Fever, rigors and vomiting | Coagulase negative staphylococci | Yes |
| 2 | 2009 | 2 | Probable | Fever and rigors | Staphylococcus epidermidis | Yes |
| 3 | 2010 | 2 | Certain | Fever, rigors, hypotension and swollen face | Streptococcus dysgalactiae | Yes |
| 4 | 2010 | 1 | Possible | Fever and dyspnoea | Staphylococcus warneri | Yes |
| 5 | 2010 | 1 | Probable | Fever, rigors and tachycardia | Acinetobacter ursingii | Yes |
| 6 | 2011 | 2 | Probable | Fever, rigors, hypertension, tachycardia, dyspnoea and vomiting | Salmonella group B | Yes |
| 7 | 2012 | 2 | Probable | Fever, rigors, tachycardia, dyspnoea, cyanosis and vomiting | Hemolytic streptococcus group C | Yes |
| 8 | 2013 | 2 | Possible | Fever, dyspnoea, vomiting and collapse | Staphylococcus hominis | Yes |
| 9 | 2014 | 4 | Certain‡ | Fever, rigors and chest pain | Staphylococcus aureus | Yes |
| 10 | 2014 | 2 | Possible | Rigors | Staphylococcus epidermidis | Yes |
| 11 | 2015 | 3 | Possible | Fever and vomiting | Enterococcus faecalis | Yes |
| 12 | 2016 | 3 | Possible | Fever, rigors, dyspnoea and chest pain | Staphylococcus epidermidis | Yes |
| 13 | 2016 | 2 | Probable | Fever, rigors, dyspnoea, hypertension, tachycardia and chest pain | Streptococcus dysgalactiae | Yes |
| 14 | 2016 | 2 | Possible | Fever, rigors, hypotension, tachycardia and decreased O2 saturation | Staphylococcus epidermidis | Yes |
| 15 | 2017 | 2 | Possible | Fever, rigors, hypertension and tachycardia | Escherichia coli | Yes |
| 16 | 2019 | 2 | Probable | Fever, rigors, dyspnoea, hypertension and tachycardia | Staphylococcus aureus | Yes |
†, a sample was taken after transfusion from the platelet bag and was cultured. The culture result was compared with the patient blood culture result. ‡, case described in 2014 hemovigilance report as ‘probable to certain’ transmission, based on genotyping. Identical species were found in patient blood culture, culture from remnant of transfused component and nasal swab from 1 of the 5 donors, but not in the associated red blood cell concentrate. TTBI, transfusion transmitted bacterial infections.
Importantly, we examined if the bacterial screening with BacT/ALERT® had become positive after release of the TTBI-associated blood products, since our blood products are released as negative-to-date. In all 16 TTBI cases, the BacT/ALERT® remained negative during the 7-day incubation period.
Positive bacterial screening with BacT/ALERT®
In 2013–2019, all pooled BC-derived PC (n=400,433) and apheresis PC (n=31,872) were cultured using the BacT/ALERT® culturing system (Table 2). A total of 1,300 pooled BC-derived PC (0.32%) and 82 apheresis PC (0.26%) were confirmed positive in the bacterial screening. For BC derived PC, both aerobic and anaerobic bottles were positive in only 7% of all positive screens, only the anaerobic was positive in 77% and only the aerobic bottle was positive in 14%. The findings were similar for apheresis platelets: in 7% of cases both bottles were positive, in 74% of cases only the anaerobic bottle and in 18% only the aerobic bottle.
Table 2
| Blood product | Total products cultured | Confirmed positive bacterial screen | Positive units that had been transfused | Transfusion reactions in recipients of positive units | ||||
|---|---|---|---|---|---|---|---|---|
| Total positive (% of products cultured) | Aerobic and anaerobic bottle | Aerobic bottle only | Anaerobic bottle only | Data about type of bottle missing | ||||
| BC derived PC | 400,433 | 1,300 (0.32%) | 88 | 177 | 995 | 40 | 439 | 20 |
| Apheresis platelets | 31,872 | 82 (0.26%) | 6 | 15 | 61 | 0 | 30 | 0 |
BC, buffy coat; PC, platelet concentrate.
Since products are released on a “negative-to-date” basis, a portion of products that were positive had already been distributed and transfused to a patient: 439/1,300 BC derived PC and 30 units/82 apheresis platelets. For the BC derived PC, a transfusion reaction was reported to Sanquin Blood Bank in only 20/439 cases. For the apheresis platelets, no transfusion reactions were reported in these cases.
Cases with positive bacterial screening after transfusion and a transfusion reaction
To examine the significance of the positive bacterial screening results in cases with transfusion reactions, we obtained clinical and microbiological data from TrackWise EQMS for the 20 cases with a positive bacterial screening after transfusion and a transfusion reaction (Table 3). Importantly, in none of these cases the same bacterial species was isolated from the blood of the patient and from the BC derived PC. Thus, none of these cases meet the criteria for TTBI. Furthermore, in none of the cases samples were taken from the PC bag after transfusion, suggesting that there was no clinical suspicion of TTBI directly after transfusion, when the unit might have been sampled for culture. At the time of recall sampling was not possible anymore.
Table 3
| Case number | Year | Severity grade | Imputability | Symptoms/diagnosis | Culture in PC sample | Patient blood culture |
|---|---|---|---|---|---|---|
| 1 | 2013 | 4 | Unlikely | Fever in patient with perforated diverticulitis and multi-organ failure | Cutibacterium spp. | Unknown |
| 2 | 2013 | 1 | Unlikely | Fever in patient with pneumonia after stem cell transplantation | Cutibacterium spp. | Negative |
| 3 | 2013 | 4 | Unlikely | Sepsis in patient with complicated abdominal surgery and peritonitis | Cutibacterium spp. | Unknown |
| 4 | 2014 | 4 | Unlikely | Clinical decline in patient with complicated cardiac surgery and multi-organ failure | Staphylococcus epidermidis | Pseudomonas spp. |
| 5 | 2014 | 4 | Unlikely | Clinical decline in patient with complicated pancreatitis and multi-organ failure | Cutibacterium spp. | Morganella spp. |
| 6 | 2015 | 1 | Unlikely | Fever (present before transfusion) without other symptoms | Cutibacterium spp. | Unknown |
| 7 | 2015 | 1 | Possible | Fever without other symptoms | Cutibacterium spp. | Unknown |
| 8 | 2015 | 1 | Unlikely | Fever (present before transfusion) without other symptoms | Cutibacterium spp. | Unknown |
| 9 | 2016 | 1 | Unlikely | Fever in AML patient treated with antibiotics before transfusion because of E. Coli infection | Staphylococcus saccharolyticus | Candida spp. |
| 10 | 2016 | 1 | Unlikely | Fever (present before transfusion) without other symptoms | Cutibacterium spp. | Grampositive cocci |
| 11 | 2016 | 1 | Unlikely | Fever without other symptoms | Cutibacterium spp. | Staphylococcus aureus |
| 12 | 2016 | 4 | Possible | Shock after vascular surgery | Hemolytic streptococcus group G | Negative |
| 13 | 2016 | 1 | Unlikely | Fever without other symptoms | Cutibacterium spp. | Staphylococcus epidermidis & Staphylococcus haemolyticus |
| 14 | 2017 | 1 | Unlikely | Fever (present before transfusion) without other symptoms | Staphylococcus saccharolyticus | Unknown |
| 15 | 2017 | 1 | Possible | Fever without other symptoms | Cutibacterium spp. | Unknown |
| 16 | 2018 | 1 | Possible | Fever and CRP raise in patient after vascular surgery | Cutibacterium spp. | Unknown |
| 17 | 2019 | 1 | Unlikely | Urticaria without fever | Cutibacterium granulosum | Unknown |
| 18 | 2019 | 1 | Possible | Fever without other symptoms | Cutibacterium spp. | Unknown |
| 19 | 2019 | 1 | Unlikely | Fever and rigors, as seen after previous transfusions | Grampositive rods | Unknown |
| 20 | 2019 | 1 | Unlikely | Fever without other symptoms | Cutibacterium spp. | Enterobacterales |
PC, platelet concentrate; AML, acute myeloid leukaemia; CRP, C-reactive protein.
In 5 out of 20 cases with a positive bacterial screen and a transfusion reaction, the transfusion reaction was potentially related to a contaminated BC-derived PC. In one case, the bacterial screening was positive with hemolytic streptococci group G. This patient was transfused with red blood cells, platelets and plasma because of bleeding during vascular surgery, and shock was reported. The bacterial screening became positive after only 0.5 days. The patient’s blood cultures remained negative. The patient did not recover and died a few weeks after the transfusion. The transfusion reaction was classified by the blood establishment as possibly related to contaminated platelets with hemolytic streptococci group G. This transfusion reaction was reported to TRIP as bacterial contamination of blood component. Following review by the Expert Committee an additional category of other reaction was recorded: subgroup of nonconfirmed sepsis, severity grade 4, imputability probable (12). In 4 cases, bacterial screening was positive with Cutibacterium spp., no positive blood cultures of the patients were reported by the hospitals, and the imputability was classified as possible, based on clinical information, even though TTBI caused by Cutibacterium spp. have only sporadically been reported (13).
In 15 out of 20 cases, imputability was unlikely. In 5 of these cases, highly virulent pathogens were cultured from the blood of the patients (Pseudomonas aeruginosa, Morganella spp., S. aureus, Enterobacterales, Candida spp.), which likely were the causative agents of the clinical presentation and the bacteria with relatively low pathogenicity in the bacterial screening seemed unrelated to the transfusion reaction. In two cases, the bacterial screening showed Cutibacterium spp. and the blood culture became positive with Gram positive cocci (no further identification possible) and coagulase negative staphylococci, respectively. In 8 cases, bacterial screening showed bacteria with limited pathogenicity (mostly Cutibacterium spp.), no positive blood cultures of the patients were reported by the hospitals and imputability of the transfusion reaction to the contaminated PC was unlikely, based on clinical information.
Reactions reported to TRIP with a positive culture of the platelet unit in the hospital
Table 4 summarizes 48 reports to TRIP in 2008–2019, other than TTBI cases, where a bacterial species was cultured in the remnant of the transfused platelet concentrate by the hospital. For all cases bacterial screening remained negative. In the absence of a positive patient blood culture assessment of potential TTBI is not applicable. All five serious reactions (severity grade ≥2 and imputability certain, probable or possible) were reviewed at case level. One case (reported as “other reaction”) was reported with clinical features suggestive of bacteremia/sepsis (fever, rigors, dyspnoea, desaturation, tachypnoea, hypotension, flank pain, nausea, diarrhoea). The culture (in the hospital) of the remnant of the blood component was positive for Escherichia coli. Blood cultures taken from the patient at the time of the reaction were negative. Therefore, this case is considered as a potential septic reaction with false negative bacterial screening and false negative patient blood culture. The symptoms of the two severe anaphylactic reactions were clinically regarded as consistent with an allergic mechanism. The other two severe cases classified as other reactions were associated with arrhythmia and hypotension in a normothermic patient and dyspnoea in a patient with bilateral pneumonia receiving treatment with antibiotics. Thus, only one additional case of potential septic transfusion reaction was identified by the review of reported cases with a positive culture of the platelet unit in the hospital.
Table 4
| Reaction | Total number of reports | Reports of grade 2 or higher† |
|---|---|---|
| Anaphylactic transfusion reaction | 3 | 2 |
| Other allergic reaction | 6 | |
| Mild (nonhemolytic) febrile reaction | 2 | |
| Nonhemolytic transfusion reaction (NHTR) | 13 | |
| Other transfusion reaction | 19 | 3 |
| Post-transfusion bacteremia/sepsis | 5 | |
| Total | 48 | 5 |
†, imputability certain, probable or possible.
Characterisation of bacterial species found in bacterial screening
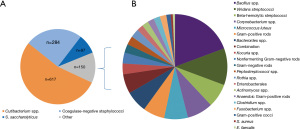
We examined the distribution of bacterial species found in our screening of pooled BC derived platelets from 2013 to 2019. As shown in Figure 1, more than half of cases of positive bacterial screening are caused by Cutibacterium spp., cultured from anaerobic bottles. Over a quarter of cases are caused by coagulase-negative staphylococci, including the anaerobic coagulase negative staphylococcus S. saccharolyticus. Figure 1 also shows the varieties of bacteria that are found occasionally.
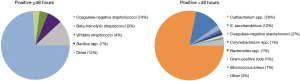
Platelets are released as negative-to-date, but are mostly administered >48 hours after production and inoculation. Therefore, we compared the bacterial species that were found in cultures turning positive within 48 hours and after 48 hours. As shown in Figure 2, the commensal and relatively slow-growing skin bacteria Cutibacterium spp. are by far the most commonly cultured bacteria after 48 hours, accounting for 78% of cases. S. saccharolyticus, an anaerobic coagulase-negative staphylococcus, accounts for 12% of cases. Before 48 hours, coagulase-negative staphylococci, also part of the commensal skin flora, are found most frequently, in 74% of cases. More virulent bacteria, including beta-hemolytic streptococci and Viridans streptococci, are usually positive in the bacterial screening within 48 hours (Figure 2). Other pathogenic bacteria, including Enterobacterales and S. aureus were seen only sporadically. We found S. aureus once, Klebsiella oxytoca once, Citrobacter freundii once, all detected before 48 hours, and Campylobacter fetus once, detected just after 48 hours (data not shown). None of these 4 PC had been administered to a patient. Together, these results show that virulent bacteria are detected only occasionally and usually within 48 hours, and thus mostly before administration.
Discussion
In this study, we examined the residual risk of septic transfusion reactions due to bacterial contamination of platelets. Based on hemovigilance data from TRIP, we identified 16 potential TTBIs [2008–2019]. Based on positive BacT/ALERT® screening data, we identified 5 cases in which contaminated platelets had possibly resulted in a transfusion reaction [2013–2019]. Including findings from both approaches, only two cases were definite TTBI, with one fatal case.
In the first approach, analysing cases of TTBI reported to the TRIP database, we found two definite TTBI cases, 6 probable cases and 8 possible cases. The definite TTBI cases involved a Streptococcus dysgalactiae and S. aureus (fatal). All these cases had a negative BacT/ALERT® screening of platelets, and thus demonstrate a small residual risk (16 cases per 668,896 distributed platelets) despite bacterial screening. Since the BacT/ALERT® remained negative during the 7-day incubation, a longer hold (later release) of platelets would not have prevented these 16 TTBI cases. However, a longer delay of sampling for culture may have prevented some cases.
After reviewing all TRIP reports, other than TTBI, where a bacterial species was cultured in the remnant of the transfused platelet concentrate by the hospital, we found one additional case that is considered a possible septic transfusion reaction with false negative patient blood culture.
In our second approach, examining all cases with positive bacterial screening of transfused platelets, we identified 5 cases in which a transfusion reaction was possibly resulting from a contaminated platelet product. The bacterial screening of the platelets showed hemolytic streptococci in 1 case and Cutibacterium spp. in the other 4 cases. There was no overlap in cases found using these two approaches.
Previously, transfusion reactions in cases with transfused, and subsequently positively screened, platelets were examined during a 2-year period [2006–2007] (14). A transfusion reaction occurred in two of 158 platelet transfusions that later became positive in the bacterial screening, but imputability of both cases was classified as unlikely. Our data from 2013 to 2019 confirms these earlier findings.
Since we found no TTBI cases resulting from transfused platelets that subsequently became positive in the bacterial screening, the value of continued incubation in the BacT/ALERT® after distribution could be questioned. Importantly, positively screened platelets that have been distributed, but have not yet been transfused, are recalled, preventing potential TTBI. Therefore, continued incubation after distribution of platelets contributes to minimizing the risk of TTBI.
In our study, we provided a detailed characterization of bacterial species found in our BacT/ALERT® screening. As in previous studies, skin bacteria, including Cutibacterium spp. and coagulase-negative staphylococci, accounted for the majority of cases (3,9). This distribution of bacterial species in the bacterial screening is highly similar to the distribution of bacterial species on the forearm and antecubital fossa (15), consistent with the skin of the donor as the main origin of bacterial contamination of PC (16). Whereas Cutibacterium spp. are mostly considered low pathogenic bacteria, they have emerged as pathogens of healthcare-associated infection, mostly in association with foreign devices, including prosthetic valve endocarditis, cardiac implantable electronic device infections and prosthetic joint infections (17). However, in the context of transfusion, the pathogenic role of these bacteria still appears low. In contrast to the frequent finding of Cutibacterium spp. in bacterial screening, only a few cases of transfusion-related sepsis due to Cutibacterium spp. have been reported (13). This may be due to low pathogenicity in combination with low bacterial load. Cutibacterium spp. do not proliferate under platelet storage condition and thus do not reach clinically significant bacterial loads (18). Virulent bacteria and high bacterial loads are associated with more-severe transfusion reactions (19).
In addition to the overall distribution of bacterial species in the screening, we also compared the distribution of species that became positive in the BacT/ALERT® before and after 48 hours. Platelets are only sporadically transfused within 48 hours from production, but release of platelets as negative-to-date poses a potential risk. As expected, but reassuring, the majority of positive screening results after 48 hours showed Cutibacterium spp. Virulent bacteria, including hemolytic streptococci, were mostly cultured before 48 hours. Interestingly, hemolytic streptococci were responsible for one certain TTBI case, for one probable TTBI case, as well for one case with a transfusion reaction possibly associated with the subsequently positive screen. These cases emphasize the important finding that these species were mostly detected within 48 hours (Figure 2).
S. aureus was detected in our bacterial screening in 1 case in 2013–2019, within 48 hours, and the product had not been transfused. In 2008–2012, S. aureus was detected in the bacterial screening in 6 cases, and in all cases the bacterial screening also became positive within 48 hours and blood products had not been transfused (unpublished data). S. aureus was responsible for one probable and one definite, fatal, TTBI during 2008–2019. The BacT/ALERT® remained negative in these cases. In the United Kingdom, S. aureus failed to be detected by the BacT/ALERT® in 4 cases during screening of 1,239,029 platelets [2011–2015], of which 3 were not transfused due to visual detection of clumps in the platelet bag (9). These findings emphasize the importance of visual inspection before transfusion as a final strategy to decrease the risk of TTBI.
The BacT/ALERT® remained negative in all TTBI cases. The number of false negatives may be reduced by later sampling of the product, first allowing proliferation of bacteria to a level above the limit of detection. Such late sampling is recommended by the FDA guideline and nowadays common practice in the United Kingdom (8,9). We have not postponed the time of sampling because this is difficult to introduce due to logistic challenges, and because our current numbers of TTBI are low. Later sampling may result in release of older products, with potentially lower quality (20), as well as more outdating. The other parameters recommended by the FDA guideline, including large volume sampling in two bottles, have been standard practice in the Netherlands since the introduction of bacterial screening in 2001 (6).
The 48 cases where hospital culture of the remnant of the PC yielded a bacterial species should theoretically also be considered as false negative BacT/ALERT® screening results. However, the majority of the transfusion reactions which led to culturing of the bag were not clinically diagnosed as cases of sepsis caused by the cultured bacteria and in no case a patient culture was positive for the same species. Obtaining samples from the remnant of the PC for culture is not standardised in most hospitals and is likely to be prone to contamination (21).
As an alternative approach to reduce occurrence of TTBI, various pathogen reduction technologies for platelets have been studies (22-24). These techniques use ultraviolet light to target nucleic acids without destroying platelet membranes, thereby preventing proliferation of bacteria. Limitations of this strategy include breakthrough of fast-growing and spore-forming bacteria and potential decrease in the quality of platelets (25-27).
To conclude, recognizing post-transfusion sepsis by clinicians, obtaining blood cultures from the patient as well as cultures of the PC are required to identify a TTBI. Therefore, adequate follow-up in case of suspicion of post-transfusion sepsis and reporting reactions to the platelet supplier and hemovigilance office are crucial. Combining information from these sources, we identified a small remaining risk of TTBI due to platelets contamination despite intervention strategies.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Sandra Ramirez-Arcos) for the series “Bacterial Contamination of Platelet Components” published in Annals of Blood. The article has undergone external peer review.
Peer Review File: Available at https://aob.amegroups.com/article/view/10.21037/aob-21-26/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://aob.amegroups.com/article/view/10.21037/aob-21-26/coif). The series “Bacterial Contamination of Platelet Components” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- FDA Center for Biologics Evaluation and Research. Fatalities reported to FDA following blood collection and transfusion: annual summary for fiscal year 2018. Available online: https://www.fda.gov/vaccines-blood-biologics/report-problem-center-biologics-evaluation-research/transfusiondonation-fatalities
- Brecher ME, Hay SN. Bacterial contamination of blood components. Clin Microbiol Rev 2005;18:195-204. [Crossref] [PubMed]
- de Korte D, Curvers J, de Kort WL, et al. Effects of skin disinfection method, deviation bag, and bacterial screening on clinical safety of platelet transfusions in the Netherlands. Transfusion 2006;46:476-85. [Crossref] [PubMed]
- Eder AF, Kennedy JM, Dy BA, et al. Limiting and detecting bacterial contamination of apheresis platelets: inlet-line diversion and increased culture volume improve component safety. Transfusion 2009;49:1554-63. [Crossref] [PubMed]
- Ramirez-Arcos S, Jenkins C, Dion J, et al. Canadian experience with detection of bacterial contamination in apheresis platelets. Transfusion 2007;47:421-9. [Crossref] [PubMed]
- de Korte D. 10 Years Experience with Bacterial Screening of Platelet Concentrates in the Netherlands. Transfus Med Hemother 2011;38:251-4. [Crossref] [PubMed]
- Benjamin RJ, McDonald CPISBT Transfusion Transmitted Infectious Disease Bacterial Workgroup. The international experience of bacterial screen testing of platelet components with an automated microbial detection system: a need for consensus testing and reporting guidelines. Transfus Med Rev 2014;28:61-71. [Crossref] [PubMed]
- United States Department of Health and Human Services Food and Drug Administration. Bacterial Risk Control Strategies for Blood Collection Establishments and Transfusion Services to Enhance the Safety and Availability of Platelets for Transfusion. 2020. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/bacterial-risk-control-strategies-blood-collection-establishments-and-transfusion-services-enhance
- McDonald C, Allen J, Brailsford S, et al. Bacterial screening of platelet components by National Health Service Blood and Transplant, an effective risk reduction measure. Transfusion 2017;57:1122-31. [Crossref] [PubMed]
- Transfusion and Transplantation Reactions in Patients (TRIP) Foundation. Trip Report 2018 Hemovigilance Extended version. Available online: https://www.tripnet.nl/wp-content/uploads/2020/08/Trip.HEMO_uitgebreid_ENGdef2020-4.pdf
- Transfusion and Transplantation Reactions in Patients (TRIP) Foundation. Trip Reports Hemovigilance Extended version, 2008-2019. Available online: https://www.tripnet.nl/en/publications/trip-reports/
- Transfusion and Transplantation Reactions in Patients (TRIP) Foundation. Trip Report 2016 Hemovigilance Extended version. Available online: https://www.tripnet.nl/wp-content/uploads/2018/05/Trip.HEMO_2016_ENG_def.pdf
- Schneider T, Breviere D, Taillefer MF, et al. Bacterial contamination of platelet concentrates by Propionibacterium acnes. Transfus Clin Biol 2000;7:540-6. [Crossref] [PubMed]
- Koopman MM, van't Ende E, Lieshout-Krikke R, et al. Bacterial screening of platelet concentrates: results of 2 years active surveillance of transfused positive cultured units released as negative to date. Vox Sang 2009;97:355-7. [Crossref] [PubMed]
- Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol 2018;16:143-55. [Crossref] [PubMed]
- Rood IG, de Korte D, Savelkoul PH, et al. Molecular relatedness of Propionibacterium species isolated from blood products and on the skin of blood donors. Transfusion 2011;51:2118-24. [Crossref] [PubMed]
- Achermann Y, Goldstein EJ, Coenye T, et al. Propionibacterium acnes: from commensal to opportunistic biofilm-associated implant pathogen. Clin Microbiol Rev 2014;27:419-40. [Crossref] [PubMed]
- Stormer M, Kleesiek K, Dreier J. Propionibacterium acnes lacks the capability to proliferate in platelet concentrates. Vox Sang 2008;94:193-201. [Crossref] [PubMed]
- Jacobs MR, Good CE, Lazarus HM, et al. Relationship between bacterial load, species virulence, and transfusion reaction with transfusion of bacterially contaminated platelets. Clin Infect Dis 2008;46:1214-20. [Crossref] [PubMed]
- Dijkstra-Tiekstra MJ, van der Meer PF, Cardigan R, et al. Platelet concentrates from fresh or overnight-stored blood, an international study. Transfusion 2011;51:38S-44S. [Crossref] [PubMed]
- Federation Medical Specialists. Guideline Blood Transfusion Policy 2019 (The Netherlands). Available online: https://richtlijnendatabase.nl/richtlijn/bloedtransfusiebeleid
- Irsch J, Lin L. Pathogen Inactivation of Platelet and Plasma Blood Components for Transfusion Using the INTERCEPT Blood System. Transfus Med Hemother 2011;38:19-31. [Crossref] [PubMed]
- Ruane PH, Edrich R, Gampp D, et al. Photochemical inactivation of selected viruses and bacteria in platelet concentrates using riboflavin and light. Transfusion 2004;44:877-85. [Crossref] [PubMed]
- Seltsam A, Muller TH. UVC Irradiation for Pathogen Reduction of Platelet Concentrates and Plasma. Transfus Med Hemother 2011;38:43-54. [Crossref] [PubMed]
- Schmidt M, Hourfar MK, Sireis W, et al. Evaluation of the effectiveness of a pathogen inactivation technology against clinically relevant transfusion-transmitted bacterial strains. Transfusion 2015;55:2104-12. [Crossref] [PubMed]
- Feys HB, Van Aelst B, Compernolle V. Biomolecular Consequences of Platelet Pathogen Inactivation Methods. Transfus Med Rev 2019;33:29-34. [Crossref] [PubMed]
- McDonald CP, Bearne J, Aplin K, et al. Assessing the inactivation capabilities of two commercially available platelet component pathogen inactivation systems: effectiveness at end of shelf life. Vox Sang 2021;116:416-24. [Crossref] [PubMed]
Cite this article as: Freudenburg-de Graaf W, Spelmink S, Heijnen J, de Korte D. Transfusion transmitted bacterial infections (TTBI) involving contaminated platelet concentrates: residual risk despite intervention strategies. Ann Blood 2022;7:26.


