
Glycophorins and the MNS blood group system: a narrative review
Introduction
After the discovery of the ABO blood group system in 1900, Landsteiner and Levine searched for more human blood groups (1). In 1927, rabbits injected with human red blood cells (RBCs) produced antibodies against M (MNS1) and N (MNS2) blood group antigens (1,2). The names of these antigens came from the word “immune” and this discovery created the second blood system now known as the MNS blood group system (2). Examples of human anti-M and anti-N were reported in later years (3).
In 1947, Walsh and Montgomery reported a female patient with puerperal fever who developed an antibody recognising the S antigen (MNS3) (4). The S antigen was named after Sydney, the capital city of New South Wales, Australia. The s antigen (MNS4), antithetical to S, was described in 1951 (4,5). Two years later, the fifth antigen U (stands for universal, MNS5) in this system was discovered (6,7). Currently, 50 antigens in the MNS system are recognised by the International Society of Blood Transfusion (ISBT) Working Party (WP) on Red Cell Immunogenetics and Blood Group Terminology (RCIBGT), Table 1 (8). These antigens are carried on glycophorin A (GPA), glycophorin B (GPB) or variant glycophorins. Amongst antibodies to MNS antigens, many are regarded as clinically significant with reported cases of haemolytic disease of the fetus and newborn (HDFN) and haemolytic transfusion reactions (HTRs). The history of the MNS system has been detailed in reference textbooks (1,2,9-11) and review articles (12-17). We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/aob-21-9).
Table 1
| n | Antigens | |
|---|---|---|
| Polymorphic antigens | n=4 | M, N, S, s |
| High-frequency antigens | n=10 | U, Ena, ENKT, ‘N’, ENEP, ENEH, ENAV, ENDA, ENEV, JENU |
| Low-frequency antigens | n=36 | He, Mia, Mc, Vw, Mur, Mg, Vr, Me, Mta, Sta, Ria, Cla, Nya, Hut, Hil, Mv, Far, sD, Mit, Dantu, Hop, Nob, Or, DANE, TSEN, MINY, MUT, SAT, ERIK, Osa, HAG, MARS, MNTD, SARA, KIPP, SUMI |
GPA, GPB, and glycophorin E (GPE)
GYPA and GYPB genes encode GPA, 150 amino acids, and GPB, 91 amino acids, respectively. GYPA and GYPB Exon 1 up to the 5' end of Exon 2 encode a 19-amino acid leader sequence, from the 3' end of Exon 2 up to Exon 4 encode amino acids residing in the extracellular region, Exon 5 in the transmembrane and Exon 6–7 in the intracellular domain. The leader sequence is cleaved after the protein is inserted into the cell membrane (1). Therefore, the mature GPA and GPB molecule has 131 and 72 amino acid residues, respectively, Figure 1 (10). GYPE encodes GPE which is a 78-amino acid molecule that includes a 19-residue leader sequence (18). The mRNA transcript produced by GYPE was shown to be very unstable affecting GPE expression (18). GPE has not been detected on the surface of RBC. However if expressed, GPE would be a 59-amino-acid molecule carrying an M antigen. The Exon 2 sequence coding for M antigen in GYPE is identical to GYPA (14,19).

GPA and GPB are single-pass transmembrane sialoglycoproteins heavily glycosylated with abundant O-glycans (10). Only GPA carries N-glycan (10). These carbohydrate molecules contribute a strong net negative charge to the surface of RBCs preventing RBC aggregation thus maintaining blood flow in the circulation (10). It is estimated that there are 1×106 copies of GPA and 2.5×105 copies of GPB per RBC (15,20).
Genes of the MNS blood group system
GYPA, GYPB and GYPE genes form a 350-kb gene cluster on chromosome 4q31.21, Figure 2A (21,22). Analysis of this region suggests that GYPA was the ancestral gene and that a series of molecular events formed GYPB and GYPE genes (23). Firstly, ancestral GYPA is duplicated. Two chromatid strands, each carrying a duplicated GYPA, misaligned. This was followed by unequal crossing-over occurring within the Alu sequences present in each strand producing a progenitor GYPB/GYPE genomic segment (23). Subsequent duplication of this segment gave rise to independent GYPB and GYPE genes. The 3' sequences for GYPB and GYPE were acquired from an unrelated genomic segment (23).
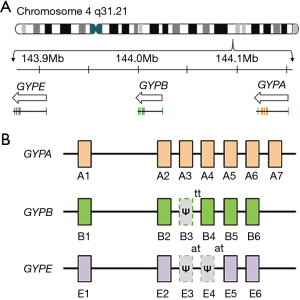
GYPA, GYPB and GYPE genes show a high degree of homology, over 95%, from the 5' flanking sequences to the Alu sequence located in Intron 5 which is approximately 1 kb downstream from Exon 5 (21,23). Sequence homology and the intron-exon gene structure organisation (Figure 2B) is thought to facilitate the numerous gene recombination events occurring in these three glycophorin genes (14). Furthermore, an approximately 1 kb region was identified as a major recombination hotspot spanning between Intron 2–Exon 3 junction and Intron 3–Exon 4 junction (10). The presence of multiple direct repeat sequences and palindromic sequences, particularly the 35-bp complex palindrome within Exon 3 of GYPA and pseudoexon 3 of GYPB, are distinct features of these genes (10). Reference sequences associated with these genes are listed in Table 2 (8).
Table 2
| Source | GYPA | GYPB | GYPE |
|---|---|---|---|
| Genomic | NG_007470.3 | NG_007483.3 | NG_009173.1 |
| Transcript | NM_002099.8 | NM_002100.6 | NM_002102.4 |
| Protein | NP_002090.4 | NP_002091.4 | NP_941391.2 |
Variant glycophorins: genetic mechanisms
Several genetic mechanisms contribute to the diversity of variant glycophorins (10,12). Unequal crossing-over and gene conversion are the two main mechanisms forming hybrid glycophorin variants (10,12). Currently, there are over 30 hybrid genes in the MNS system (8).Examples of these hybrid alleles are shown in Table 3. The third mechanism is single nucleotide polymorphism (10,12).
Table 3
| Mechanisms | Variant alleles |
|---|---|
| Unequal crossing-over | |
| GYP(A-B) | e.g., GYP*Hil, GYP*JL |
| GYP(B-A) | e.g., GYP*Dantu, GYP*Sch, GYP*MOT |
| Gene conversion | |
| GYP(A-B-A) | e.g., GYP*Vw, GYP*Hut, GYP*Zan |
| GYP(B-A-B) | e.g., GYP*Mur, GYP*Bun, GYP*Hop, GYP*HF, GYP*Kip |
| GYP(A-E-A) | e.g., GYP*Mar |
| GYP(B-E-B) | e.g., GYP*Man, GYP*Ros, GYP*Dia, GYPB-E(2-4)-B |
| Single nucleotide polymorphism causing missense mutations or splicing variants | |
| GYPA c.91A>C | e.g., GYPA*SUMI |
| GYPB c.161G>A | e.g., GYPB*Mit |
| GYPA c.232G>A | e.g., GYPA*Erik or GYP*EBH |
Unequal crossing-over
In this mechanism, GYPA and GYPB genes misaligns during meiosis (13). The two sister chromatid strands exchange genetic material of unequal length generating two hybrid genes in reciprocal arrangements. As a result, one strand received less (Lepore type) and the other received more (anti-Lepore type) genetic material than what each initially gave (14). Unequal crossing-over forms GYP(A-B) and GYP(B-A) hybrid alleles, Table 3.
Gene conversion
This mechanism also occurs during meiosis when nucleotide sequences from a donor chromatid strand replace a homologous sequence in the acceptor chromatid strand (10). This transfer of genetic material is non-reciprocal. Gene conversion between GYPA and GYPB forms GYP(A-B-A) and GYP(B-A-B) hybrid genes (13,14). Examples of GYP(B-A-B) hybrid genes are formed when the defective donor splice site “tt” in GYPB pseudoexon 3 is replaced by the functional splice site “gt” from GYPA Exon 3 (10), Table 3. Conversely, an active splice site on GYPA Exon 3 replaced by the inactive splice site from GYPB pseudoexon 3 produces a variant glycophorin called GP.Zan (26). GYPE also recombines with GYPA forming GYP(A-E-A)—encoding GP.Mar—and with GYPB to form GYP(B-E-B) (24,25). Few examples of GYP(B-E-B) alleles have been identified and they differ from each other based on the position and length of the GYPE gene insert, Table 3 (24,25).
Recently, a trypsin-resistant M antigen was identified in 0.05% of the Japanese population (24). This antigen was encoded by a novel variant glycophorin GYPB-E(2-4)-B (24). Exons 3 and 4 of this allele are pseudoexons. The final protein is a 59-amino-acid GP(E-B) molecule expressing M antigen. To our knowledge, this was the first report of a glycophorin molecule expressing a GYPE product. GYPB-E(2-4)-B has a similar structure to GYPB-E-B.Ros allele reported by Willemetz et al. in a Caucasian individual from Portugal (25). However, the 5' gene breakpoint for GYPB-E-B.Ros has not been fully defined (24,25). Two other examples of GYPB-E-B alleles were identified in African individuals—GYPB-E-B.Man (Gambia) and GYPB-E-B.Dia (Mali) (25).
Single nucleotide variants (SNVs)
SNVs in the exon or intron regions of GYPA and GYPB genes produce variant glycophorins either by an amino acid change, or disrupting the normal splicing mechanisms if the nucleotide is adjacent to or near the splice site, Table 3. Splice sites are important markers during splicing, when introns are removed and exons are fused together, by spliceosomes. Single base substitutions in the donor splice site (gt) will cause skipping of the preceding exon (27). Examples of glycophorin variants formed by this mechanism include GP.EBH and GP(P2).
- SNV in the exon—the GP.EBH phenotype arose from a c.232G>A (p.Gly59Arg) in GYPA Exon 3. This SNV is located adjacent to the “gt” splice site in Intron 3 and produces several transcripts. One transcript forms GPA carrying the ERIK antigen (p.Gly59Arg). Another forms a GPA molecule, lacking the Exon 3 product, expressing Sta (MNS15) antigen (28).
- SNV in the intron—the GYPB*P2 allele has a c.270+5G>T polymorphism located in Intron 5. This base change results in skipping of Exon 5 forming GP(P2) expressing a S–s–U+var phenotype (29).
Loss of GYPB gene (S–s–U– phenotype)
Homozygous deletion of glycophorin genes generate null phenotypes such as MkMk (deletion of GYPA and GYPB), En(a–) (a deletion of GYPA) and S–s–U– (deletion of GYPB) (8). The S–s–U– phenotype is present in approximately 1% of individuals of African heritage and the predominant GYPB deletion alleles have been identified (30-32). However, the major alleles responsible for the S–s–U– in Asians, Caucasians and other population groups are yet to be determined.
S–s–U– phenotype in Africans
Studies by Leffler et al. (32), Gassner et al. (30), and Lane et al. (31) identified several examples of GYPB deletion alleles (whole and partial gene deletion) that give rise to S–s–U– phenotype (Table 4). The two most common alleles identified in these studies were GYPB*05N.01 and GYPB*05N.02 alleles (30-32). Both alleles have a deletion span of over 100 kb in the GYP locus which includes the entire GYPB gene. These were observed in individuals from West Africa (The Gambia, Sierra Leone, Nigeria, Burkina Faso and Cameroon) and East Africa (Tanzania and Kenya) (30,32). In addition, GYPB*05N.01 was identified in a sample from North Africa (Algeria), Central Africa (DR Congo) and Southern Africa (South Africa) (30). A 19-kb deletion within the GYPB gene, GYPB*05N.03, was identified in an African Barbadian individual (31).
Table 4
| ISBT allele name | GYP deletion size described by Gassner et al. (30) [2020] | Gassner et al. (30) [2020] | Lane et al. (31) [2020] | Leffler et al. (32) [2017] |
|---|---|---|---|---|
| GYPB*05N.01 | 110.24 kb deletion includes the entire GYPB | 110-kb deletion | DEL_B_LEFT | DEL1 |
| GYPB*05N.02 | 103.26 kb deletion includes the entire GYPB | 103-kb deletion | DEL_B_RIGHT | DEL2 |
| GYPB*05N.03 | 18.61 kb deletion includes GYPB exon 2–6 | 19-kb deletion | DEL_PART_B | DEL8 |
Based on the similarity in size and location of the deletion within the GYP locus, the authors of this manuscript assigned DEL_PART_B allele as GYPB*05N.03. ISBT, International Society of Blood Transfusion.
S–s–U– phenotype in Asians
Rare examples of S–s–U– have been reported in Asians (33). In 1972, a pregnant woman of Indian heritage was typed as U– (33). She had post-partum transfusion after her first pregnancy. In her third pregnancy, she delivered a U+ baby showing signs of mild HDFN (33). Anti-U and anti-c antibodies were detected in the mother and were eluted from the baby’s RBCs. The mother has two siblings who were also S–s–U– (33). Another case of HDFN due to anti-D and anti-U antibodies in a pregnant D–, S–s–U– woman in India was described (34).
Three GYPB deletion types were identified in Asian individuals: (I) GYPB*05N.03 in a Bengali individual (31,32), (II) a 112-kb deletion which includes GYPA Exons 4–7 to GYPB Exon 1 identified in a Gujarati Indian (31), and (III) a 224-kb deletion, DEL_EB-1c, in the GYP locus that includes the whole GYPB and GYPE genes in a Sri Lankan Tamil individual (31,32). Three DEL_EB types described by Lane et al. (31) resembles the DEL6 variant identified by Leffler et al. (32). The predominant GYPB deletion allele in Asians is not known.
S–s–U– phenotype in Caucasians
The S–s–U– phenotype was reported in a Caucasian blood donor during routine phenotyping (35). The propositus was identified as “Fav.”. Three family members of “Fav.” were also S–s–U– (35). DNA analysis for “Fav.” showed that GYPB Exon 2–5 and GYPE Exon 1 were deleted (18). This allele is designated as GYPB*01N (18,31,35). GYPB*01N allele was not described in the cohort of African and Asian population groups in recently published studies (30-32). Other examples of S–s–U– in Caucasians (Finland) have been reported (1).
S–s–U– phenotype in the Americas
Lane et al. identified two other 224-kb GYPB deletion types: DEL_EB-1a in an African from Barbados and DEL_EB-1b in a Peruvian individual (31). S–s– phenotype was found in two Central American Indians (Honduras) (1).
MNS antigens recently recognised by ISBT
Review articles on the MNS system published before 2014 included 46 blood group antigens. Since then, four new blood group antigens have been added (1,2,13,14).
SARA (MNS47)
A regular blood donor whose cells were used as a reagent RBC for antibody identification reacted to a serum from a patient (36). Serological studies showed the antigen is novel and inherited (36). This antigen was originally named “SARAH” but is now called SARA (8,36). A whole-exome sequencing study on SARA+ individuals, from two independent families, showed that GYPA c.240C>T was the genetic basis for the SARA antigen (37). In 2015, the ISBT WP on RCIBGT assigned SARA as MNS47 (37). At least two cases of HDFN due to antibodies against SARA antigen have been reported (38,39).
KIPP (MNS48)
RBCs from a blood donor of German origin showed a unique reactivity profile using antibodies with known specificity to low-frequency MNS antigens (40). This phenotype was called GP.Kip (40). A laboratory report (J. Poole, International Blood Group Reference Laboratory, personal communication, 27 October 1988) stated that ‘Kip’ is the short form for the name Kippenhahn, the German propositus. GP.Kip was also described in an Australian blood donor (40). DNA sequencing for the Australian example revealed GYP*Kip as a hybrid GYP(B-A-B) gene (41). GP.Kip is Mia+ and carries p.Ser51 which is distinct from other Mia+ GYP(B-A-B) hybrid glycophorins (41). Several Japanese individuals have been identified as GP.Kip (42-44). The KIPP antigen on GP.Kip is recognised by two anti-Hop(+Nob) antisera, Anek and Raddon (40). Another Mia+ hybrid glycophorin called GP.MOT also express KIPP antigen (45,46).
JENU (MNS49)
A Thai individual with thalassemia was transfused with RBCs (47). Following transfusion, anti-E, anti-c, anti-Jkb, anti-S and an antibody to a high-frequency antigen on GPB were identified in the patient’s serum (47). Epitope mapping analysis using 12-mer peptides, representing the extracellular domain of GPB, showed that an antibody in the patient’s plasma recognised an epitope with the sequence SYISSQTNGETG (47). This sequence is encoded by GYPB Exon 2 and Exon 4 producing 38SYISSQTN45 and 46GETG49, respectively. This epitope is called JENU (47). The name JENU is a combination of ‘JE’—the first two letters from surname of the antibody producer, and ‘NU’ from the high-frequency antigens ‘N’ (MNS30) and U (MNS5) on GPB. Phenotyping and genotyping showed the patient was GP.Mur homozygote (GP.Mur/GP.Mur) (47). GP.Mur/GP.Mur individuals do not express normal GPB and are, therefore, JENU-negative (47). These individuals can produce antibodies to GPB including anti-JENU (47,48).
SUMI (MNS50)
A patient’s serum was found reactive to RBCs from a blood donor during compatibility testing but non-reactive to the antibody identification panel of RBCs (49). Subsequent serological investigations were performed and called this antigen—SUMI. An anti-SUMI monoclonal-antibody producing cell line was created. Anti-SUMI was used to screen 541,522 blood donors and identified 23 were SUMI-positive (49). Molecular analysis showed all 23 individuals carried a single nucleotide change GYPA c.91A>C (p.Thr31Pro). SUMI is a low-frequency antigen on GPA with a prevalence of 0.0042% in blood donors in Japan (49). SUMI antigen was designated as MNS50 by the ISBT WP on RCIBGT.
Mia and its associated hybrid glycophorins
Mia (MNS7) is immunogenic and its clinical significance is widely reported (16). Currently, eight hybrid glycophorins express Mia, Table 5. A new Mia+ hybrid glycophorin called GP.MOT was recently described in a Japanese blood donor (45,46). GP.MOT is encoded by GYP*MOT—a GYP(B-A) hybrid gene. GYP*MOT is formed when a section of GYPA is replaced by homologous sequences from GYPB (45,46). The resulting structure for GYP*MOT is GYPB(1-2)-BA(3)-A(4-7). GYP*MOT was previously reported as GYP(A-B-A) hybrid gene (45,46). The Exon 3 sequence, predicting amino acid sequence DKHKRDTYPAHTANEVSEISVTTVSPPEEET, for GYP*MOT is identical to GYP*Kip (41,46). Both GP.MOT and GP.Kip are KIPP+, Table 5.
Table 5
| GP | Mia | Vw | Hut | Mur | MUT | Hop | Hil | TSEN | MINY | KIPP |
|---|---|---|---|---|---|---|---|---|---|---|
| GP.Vw | + | + | – | – | – | – | – | – | – | – |
| GP.Hut | + | – | + | – | + | – | – | – | – | – |
| GP.Mur | + | – | – | + | + | – | + | – | + | – |
| GP.Hop | + | – | – | + | + | + | – | + | + | + |
| GP.Bun | + | – | – | + | + | + | + | – | + | + |
| GP.HF | + | – | – | – | + | – | + | – | + | – |
| GP.Kip | + | – | – | + | + | – | + | – | + | + |
| GP.MOT | + | – | NT | + | + | – | – | NT | – | + |
The Mia epitope is recognised by two murine monoclonal antibodies—CBC-172, binding to epitope 48-HKRDTYAA-55, and GAMA210, binding to epitopes 44-TNDKHKRD-51, and 43-QTNDMHKR-50 (54,55). These moAb equally gave strong agglutination reactions (3+ to 4+ in a “0–4” scale) against a panel Mia positive RBCs (56). CBC-172 and GAMA210 monoclonal antibodies have been used to screen for Mia in large blood donor populations, Table 6 (44,57-59).
Table 6
| Country | moAb used | Total tested | Mia+ donors | Frequency |
|---|---|---|---|---|
| Japan [2019] (44) | CBC-172 | 826,379 | 831 | 0.1% |
| Australia [2020] (57) | CBC-172 | 5,098 | 11 | 0.22% |
| USA [2019] (58) | GAMA210 | 4,600# | 103 | ND |
| India [2016] (59) | GAMA210 | 1,000 | 1 | 0.1% |
#, number of blood donations from Asian American blood donors. ND, not determined.
Generally, Mia is rare in Caucasian (9) and African (60,61) population groups and is more commonly found in Asian populations (62-65). In the 1960s, studies on the indigenous population in the American continents reported Mia in Seneca Indians in North America, Quecha Indians in Ecuador (66) and in a Mataco individual in Argentina (67). Of all the Mia+ hybrid glycophorins, GP.Mur is most commonly encountered while others are geographically or ethnically-specific. The availability of anti-Mia typing reagents and in combination with molecular typing allowed identification of Mia+ hybrid glycophorins in other population groups, Table 7.
Table 7
| GP | Initial reports | Current reports |
|---|---|---|
| GP.Vw | Reported in Caucasians (9) and Thais (63). One Kekchi individual from Guatemala was Vw+ (68) | One Vw+ blood donor was reported in India (69) and 17 in Japan (44) |
| GP.Hut | Previously reported in Caucasians (9) and Thais (63) | GP.Hut was described in two African American blood donors (61) and in 182 Japanese blood donors (44) |
| GP.Bun | GP.Bun is usually found in the Thai population (53,70) | GP.Bun blood donors were identified in China (65,71), the US (58), Japan (44), and Australia (57) |
| GP.Hop | Two GP.Hop blood donors were reported. One in the UK, GP.Hop (MH) (72), and the other is from Australia, GP.Hop (ES) (72,73) | Nine blood donors in Thailand were phenotyped as GP.Hop (74) |
| GP.HF | Identified in the Japanese population (44,52,75) | First report of GYP*HF in a Chinese blood donor (65) |
| GP.Kip | GP.Kip was reported in a German individual and in an Australian blood donor (40) | Several GP.Kip blood donors were identified in Japan (42,44) |
| GP.MOT | No prior reports | Japanese blood donor (45,46) |
Variant glycophorins with altered antigen expression
Qualitative expression of s (MNS4) in GP.Mur, GP.Bun and GP.Hil
The products of GYPB Exon 2 (B2) and Exon 4 (B4) form the extracellular region of GPB. The s antigen, found on the B4 segment of GPB, resides near the B2-B4 junction site (10). In s+ hybrid glycophorins, the B2-B4 junction site does not exist. This is either due to the insertion of an Exon 3 product between B2 and B4 (e.g., GP.Mur or GP.Bun) or that the B2 segment was replaced by products of GYPA (e.g., GP.Hil). Studies have shown that structural changes adjacent to B4 alter the s presentation and may not be recognised by some anti-s typing reagents, Table 8 (10,47,76,77).
Cleghorn reported that in one GP.Mur+ Chinese family, one in five potent anti-s antisera failed to react with the s/(Hil) antigen (78). This is consistent with recent studies showing GP.Mur homozygote RBCs reacted variably to a panel of anti-s typing reagents (47,76,77,79). Anti-s monoclonal antibody P3BER does not recognise the s antigen on GP.Mur, GP.Bun, and GP.Hil RBCs suggesting these hybrid glycophorins express a variant s antigen (47,76,77). Anti-s has been reported in a s+ GP.Mur individual (1).
In one study, plasma from an alloimmunised GP.Mur/GP.Mur individual was tested against synthetic 12-mer peptides representing the extracellular domain of GPB (47). Peptide mapping analysis showed three distinct reactivity domains (47). The first domain (peptides 4–7) represents the JENU epitope. The second (peptide 9) and third (peptides 11–13) domains represent epitopes for s and U, respectively (47). Data suggests that three antibodies were present: anti-JENU, anti-s, and anti-U. However, at the time of publication, only anti-JENU was reported (47).
Qualitative U (MNS5) expression in GP.Mur
GP.Mur homozygote individuals do not possess normal GPB and are at risk of alloimmunisation when exposed to RBCs carrying normal GPB (47). A case was reported in a pregnant GP.Mur/GP.Mur individual of Thai ethnicity (48). Plasma from the patient reacted positive with all routine screening panel cells. Antibody identification investigations showed that the patient’s plasma failed to reactive with S–s–U– and MkMk RBCs but were weakly positive with S–s–U+ cells identifying an anti-U antibody (48). This suggests that GP.Mur/GP.Mur individuals express a variant form of U antigen and can form anti-U antibody when exposed to normal GPB.
Altered S (MNS3) expression in GPB.Mit
The Mit (MNS24) antigen, GYPB c.161G>A, is carried on GPB.Mit (1). Mit+ RBCs are usually associated with reduced S expression (1). In one case report, an apparent alloanti-S was detected in a S+s+ male, Caucasian patient (80). The patient’s RBCs were tested using multiple anti-S reagents and consistently gave positive results. Molecular typing by SNP-microarray predicted S–s+ while DNA sequencing predicted S+s+ (80). In addition, a c.161G>A (p.Arg54His) was detected predicting Mit antigen. This is the first report demonstrating GPB.Mit RBCs express an altered S and Mit+ individuals are at risk of alloimmunisation producing alloanti-S antibody (80).
Antibodies to hybrid glycophorins in patients and blood donors
Mia+ screening cells are used to detect antibodies against antigens carried on hybrid glycophorins. Antibodies to these antigens are commonly reported in several patient groups in Asia. The incidence of anti-Mia antibodies was reported at 0.08% in 20,283 patients of Guangxi, China (81) and 2.07% in 143 thalassemia patients in Thailand (82). In Malaysia, anti-MUT, anti-Mur, and anti-Mur + MUT was detected in 0.60% (n=70,543) patients in a tertiary care hospital (83). In a study in Brazil, Nakasone et al. reported the prevalence of anti-Mia in 7,119 patients was 0.41% (84). Nakasone et al. recommended the use of Mia+ screening cells in Brazil in areas with a significant Asian population (84).
In Japan, the frequency of GP.Hil in blood donors is 0.03% (4/13,546) (44). GP.Hil and GP.JL RBCs were used to screen sera from 137,340 blood donors (44). Anti-Hil and anti-MINY antibodies were detected in 10 and 3 blood donors, respectively (44).
Cases demonstrating the clinical significance of hybrid glycophorins
Antibodies to MNS antigens are frequently naturally occurring and can be ignored unless these antibodies are reactive at 37 °C (14). There are rare examples of anti-M and anti-N antibodies reactive at 37 °C causing immediate and delayed HTR (85). Antibodies to low-frequency MNS antigens Mia, Hut, and Mur are also clinically significant and have been implicated in immediate and delayed HTR and HDFN (15,16,85). The case studies presented below are consistent with previous reports (16).
HDFN due to anti-Mur [2016]
A baby exhibited jaundice 24 hours post-delivery and was treated with phototherapy. One week later, jaundice was still evident and another round of phototherapy was given (86). The mother, father and baby were all Group A, D+ (86). Mother’s plasma failed to react with standard antibody screening cells, although reactive positive with RBCs from the baby and father. Serological investigations showed that antibody from the mother reacted with Mur+ RBCs. DNA typing showed the father was homozygous for GYP*Mur (86). Based on serological and molecular data, the authors concluded that the antibody most likely caused HDFN was anti-Mur. The ancestry of the mother is Chinese and the father is Vietnamese (86). The authors signalled that in the United States, it is prudent to consider antibodies to variant glycophorins in patients of Asian ancestry when investigating for HDFN (86).
HTR due to anti-Mur antibodies in a patient with leukemia [2017]
A 41-year-old male Hispanic individual was diagnosed with leukemia (87). The patient is transfusion-dependent requiring 1–2 units of RBCs every 1–2 weeks. After one such blood transfusion event, the patient’s haemoglobin dropped to 6.6 g/dL. Two months post-transfusion, anti-Jka and anti-Mur antibodies were detected in the patient’s plasma (87). A lookback study was undertaken to determine the ethnicity of blood donors linked to the RBCs received by the patient. Of the 30 blood donors, four had Asian ancestry (87). In regions where a significant population of blood donors are of Asian ancestry, screening for anti-Mur in chronically-transfused patients could help prevent HTR.
Suspected HTR due to anti-Mia and anti-Vw antibodies in a sickle cell disease patient [2019]
A regularly-transfused African American patient with sickle cell anemia received a unit of packed RBCs. Following-transfusion, the patient experienced severe back pain (88). Post-transfusion heart rate and blood pressure readings were higher compared to pre-transfusion (88). Pre- and post-transfusion samples from the patient did not react with an antibody screening RBC panel. Antibody screening using RBCs expressing low-prevalence antigens showed the patient had multiple antibodies: anti-Vw, anti-Mia and anti-Goa (88).
HTR due to anti-Hut in a geriatric patient [2020]
A 74-year-old female individual presented with rigors, tachycardia, and fever during transfusion. Post-transfusion sample from the patient indicated hyperbilirubinemia (89). The patient’s pre- and post-transfusion samples were negative with the reagent RBC panel and but reacted with one of two donor units of RBCs. RBC from the donor was M+ N‒ Mia+. Patient’s plasma was positive with GP.Hut (Mia+ Mur‒ Hut+ MUT+) cells and negative with GP.Mur cells (Mia+ Mur+ Hut‒ MUT+). This is the first report on anti-Hut causing HTR (89).
HDFN due to anti-Mia [2020]
The Mur antigen (MNS10), expressed by hybrid glycophorins such as GP.Mur (Table 6), is considered low-frequency in Caucasian and African populations but is more commonly found in Southeast Asian and East Asian populations (90). A clinical case involving a pregnant individual of Chinese and Filipino ethnicity delivered a baby showing signs of HDFN. The infant’s RBC were DAT positive. Serological studies showed anti-Mia was present in the maternal plasma (90). The father’s RBC were Mia+ Hil+ and MINY+. DNA sequencing analysis showed the father was GYPB/GYP*Mur. Anti-Mia antibodies are not routinely detected in North America because screening cells used in the laboratory are not Mia+. The authors advocate the use of Mia+ screening cells to improve detection of anti-Mia especially in individuals of Asian background suspected with antibodies to low-frequency antigens (90).
Genotyping for hybrid glycophorins
Hemagglutination technique is the conventional method to identify blood group antigens (91). However, serological typing has limitations. It is difficult to accurately phenotype recently transfused patients or when reliable typing reagents are not available (91). Commercial monoclonal antibody GAMA210, a typing reagent for Mia, has lately become available (58). However, characterising hybrid glycophorins requires more than one typing reagent and is performed only in specialised laboratories who have access to rare antisera. Molecular typing can overcome these challenges. A brief description of genotyping techniques used to type for hybrid glycophorins is described below. This list is non-exhaustive.
Polymerase chain reaction-sequence specific primer (PCR-SSP)
Palacajornsuk et al. designed two sets of primers to detect six hybrid glycophorin genes (92). The first primer set produces two types of amplicons depending on the gene present—148-bp band for GYP*Mur, GYP*Hop, and GYP*Bun and a 151-bp for GYP*Hut and GYP*HF. The second set of primers targets GYP*Vw producing a 296-bp band (92). A 434-bp human-growth hormone band was used an internal DNA control. While one primer set was specific for only one hybrid glycophorin, GYP*Vw, the other targets five hybrid glycophorins and would require DNA sequencing to define the specific hybrid glycophorin gene present (92).
High-resolution melting (HRM) analysis
HRM analysis is a powerful screening tool to detect polymorphisms based on the melting property of double-stranded DNA (dsDNA) (93). HRM requires a real-time PCR equipment to perform and uses an intercalating dye. This dye emits fluorescence only when bound to dsDNA (93). Fluorescence is monitored throughout the testing process. HRM is a two-step process. The first step is a standard PCR procedure. As more amplicons are generated, fluorescence is increased. The second is the HRM analysis step. At this stage, heat is gradually increased to promote denaturation of dsDNA. As amplicons dissociate, fluorescence is decreased. Fragment length, GC content, sequence and heterozygosity influence the unique DNA melting profile of amplicons (93). This signature melt profile is the basis for genotyping assignments in comparison to known DNA controls. HRM have been successfully applied to genotype for GYP*Mur, GYP*Bun, and GYP*HF and to determine zygosity for hybrid glycophorin genes (47,65).
Matrix-assisted laser desorption/ionisation, time-of-flight mass spectrometry (MALDI-TOF MS)
The MassARRAY (Agena Bioscience) system combines PCR and MALDI-TOF MS technologies to detect single nucleotide polymorphisms (94). After PCR amplification, custom-designed primers hybridise to target regions (94,95). Annealed primers are extended by a single base, mass-modified dideoxynucleotide terminators, specific to the complementary nucleotide on the template (95). Products are spotted onto a chip and then shot with a laser beam to desorb and ionise. The time it takes for the ionised molecules to travel towards the detector—TOF—is calculated. TOF is proportional to mass of the extended product (95). MALDI-TOF MS is powerful in detecting SNV alleles and hybrid glycophorin alleles (73,94). MALDI-TOF MS phenotype predictions for M/N and S/s antigens have been shown to be highly concordant with serology (94).
Multiplex ligation-dependent probe amplification (MLPA)
The MLPA genotyping platform was developed by MRC Holland, The Netherlands (96,97). This technique uses two oligonucleotide probes to interrogate a particular target sequence. One of two probes carry a unique length of nucleotide sequences making it distinct with other probes in a multiplex PCR set-up (96). Once the two adjacent probes annealed to their target sequences, ligation occurs creating a single fragment (96). The ligated probes are then amplified and PCR products are size-separated by capillary electrophoresis. MLPA analysis software converts these fragments as peaks and are analysed to predict phenotype and zygosity (96). MLPA has been evaluated for RBC genotyping and can detect several types of hybrid glycophorins including GYP*Mur (96,97).
Fluidic microarray
The ID Core XT genotyping platform (Progenika/Grifols) is a multiplex PCR, hybridisation-based assay to detect multiple alleles encoding blood group antigens belonging to 10 blood group systems (98,99). This platform uses fluorochrome-labelled microspheres coupled with an allele-specific oligonucleotide probe (98,99). DNA is amplified using nucleotides labelled with biotin. Biotin-labelled amplicons hybridises to oligonucleotide probes. Following this step, a fluorescent molecule called streptavidin-phycoerythrin is added that will bind to biotin. Fluorescence signal is then detected by Luminex flow analyser (98). Data is analysed by ID Core XT analysis software (98). ID Core XT platform targets nucleotide at GYP c.140 to predict Mia (MNS7) (99).
Next-generation sequencing (NGS)
NGS also called Massively Parallel Sequencing (MPS) is a high throughput DNA sequencing platform (91). MPS-based whole-genome sequencing (WGS) and whole-exome sequencing using Illumina HiSeq and MiSeq, respectively, have been used to predict multiple RBC antigens (100-102). Briefly in these MPS platforms, DNA is fragmented and adapters are attached to the end of the fragments in preparation for DNA amplification. After amplification, dsDNA fragments are denatured forming single-stranded templates. Sequencing by synthesis begins when primers are extended with fluorescent-labelled dNTPs. Laser is applied and the fluorescent signal is detected. MiSeq sequencing platform, in particular, can generate sequence reads up to 300 bp. Sequence reads are then aligned to the reference sequence and a variant call file is generated. Data is interpreted to predict blood group antigens. Short-read sequences produced by MPS is powerful in detecting SNVs (100-102). However, using short-read sequences to characterise large structural variants, especially those formed by RHD/RHCE genes, and GYPA/GYPB genes can be challenging (101). Hybrid alleles, exon/intron deletions, and exon/intron duplications are examples of these structural variants. Long-read sequences, over 5 kb, can overcome this limitation (103).
Discussion
The cumulative reports of new gene GYPB deletion alleles, hybrid alleles, and blood group antigens demonstrate the diversity and complexity of the MNS blood group system. Case reports of HTR and HDFN highlight the immunogenic potential and clinical significance of MNS blood group antigens. Gene conversion and unequal crossing-over are genetic mechanisms frequently associated with the MNS system. These molecular mechanisms that produce novel blood group antigens are the same genetic mechanisms that disrupt them. For example, in GP.Mur, gene conversion facilitated expression of Mia, Mur, MUT, Hil, MINY antigens but also disrupted the expression of JENU antigen. Newly reported case studies associated with antibodies to MNS antigens were an enduring reminder of the clinical significance of this system. The identification of Mia+ hybrid glycophorins in other population groups suggests these variant glycophorins may not be as geographically or ethnically exclusive as first thought. The introduction of DNA sequencing technology has allowed us to identify variant glycophorins when antisera is not available. Recent history suggests that the MNS story is not finished. With the utilisation of current and emerging typing platforms, discovery of new alleles and gene variants is almost inevitable.
Acknowledgments
We thank Candice Davison, Eunike McGowan, and Brett Wilson for reviewing this manuscript.
Funding: The Australian governments fund Australian Red Cross Lifeblood for the provision of blood, blood products and services to the Australian community.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Yann Fichou) for the series “Molecular Genetics and Genomics of Blood Group Systems” published in Annals of Blood. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/aob-21-9
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aob-21-9). The series “Molecular Genetics and Genomics of Blood Group Systems” was commissioned by the editorial office without any funding or sponsorship. RLF serves as an editorial board member of Annals of Blood. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Daniels G. Human blood groups. 3rd ed. Oxford: Wiley-Blackwell Publishing, 2013.
- Reid ME, Lomas-Francis C, Olsson ML. The blood group antigen facts book. 3rd ed. San Diego: Elsevier Academic Press, 2012.
- Farr AD. Blood group serology--the first four decades (1900--1939). Med Hist 1979;23:215-26. [Crossref] [PubMed]
- Walsh RJ, Montgomery CM. A new human iso-agglutinin subdividing the MN blood groups. Nature 1947;160:504-5. [Crossref] [PubMed]
- Sanger R, Race RR. Subdivisions of the MN blood groups in man. Nature 1947;160:505. [PubMed]
- Wiener AS, Unger LJ, Gordon EB. Fatal hemolytic transfusion reaction caused by sensitization to a new blood factor U: report of a case. J Am Med Assoc 1953;153:1444-6. [Crossref] [PubMed]
- Wiener AS, Unger LJ, Cohen L. Distribution and heredity of blood factor U. Science 1954;119:734-5. [Crossref] [PubMed]
- International Society of Blood Transfusion (ISBT). Red cell immunogenetics and blood group terminology. 2020. Available online: http://www.isbtweb.org/working-parties/red-cell-immunogenetics-and-blood-group-terminology/
- Race RR, Sanger R. Blood groups in man. 6th ed. Oxford: Blackwell Scientific Publications, 1975.
- Huang CH, Blumenfeld OO. MNS blood groups and major glycophorins: molecular basis for allelic variation. In: Cartron JP, Rouger P. editors. Molecular basis of human blood group antigens 6. New York: Plenum Press, 1995:153-88.
- Issitt PD, Anstee DJ. Applied blood group serology. 4th ed. Durham: Montgomery Scientific Publications, 1998.
- Blumenfeld OO, Huang CH. Molecular genetics of glycophorin MNS variants. Transfus Clin Biol 1997;4:357-65. [Crossref] [PubMed]
- Palacajornsuk P. Review: molecular basis of MNS blood group variants. Immunohematology 2006;22:171-82. [Crossref] [PubMed]
- Reid ME. MNS blood group system: a review. Immunohematology 2009;25:95-101. [Crossref] [PubMed]
- Lomas-Francis C. Miltenberger phenotypes are glycophorin variants: a review. ISBT Science Series 2011;6:296-301. [Crossref]
- Heathcote DJ, Carroll TE, Flower RL. Sixty years of antibodies to MNS system hybrid glycophorins: what have we learned? Transfus Med Rev 2011;25:111-24. [Crossref] [PubMed]
- Castilho L. An update on the MNS blood group system. Immunohematology 2019;35:61-2. [Crossref] [PubMed]
- Vignal A, Rahuel C, London J, et al. A novel gene member of the human glycophorin A and B gene family. Molecular cloning and expression. Eur J Biochem 1990;191:619-25. [Crossref] [PubMed]
- Rahuel C, Elouet JF, Cartron JP. Post-transcriptional regulation of the cell surface expression of glycophorins A, B, and E. J Biol Chem 1994;269:32752-8. [Crossref] [PubMed]
- Gardner B, Parsons SF, Merry AH, et al. Epitopes on sialoglycoprotein alpha: evidence for heterogeneity in the molecule. Immunology 1989;68:283-9. [PubMed]
- Kudo S, Fukuda M. Structural organization of glycophorin A and B genes: glycophorin B gene evolved by homologous recombination at Alu repeat sequences. Proc Natl Acad Sci U S A 1989;86:4619-23. [Crossref] [PubMed]
- Fukuda M. Molecular genetics of the glycophorin A gene cluster. Semin Hematol 1993;30:138-51. [PubMed]
- Onda M, Kudo S, Rearden A, et al. Identification of a precursor genomic segment that provided a sequence unique to glycophorin B and E genes. Proc Natl Acad Sci U S A 1993;90:7220-4. [Crossref] [PubMed]
- Tsuneyama H, Isa K, Watanabe-Okochi N, et al. An unusual variant glycophorin expressing protease-resistant M antigen encoded by the GYPB-E(2-4)-B hybrid gene. Vox Sang 2020;115:579-85. [Crossref] [PubMed]
- Willemetz A, Nataf J, Thonier V, et al. Gene conversion events between GYPB and GYPE abolish expression of the S and s blood group antigens. Vox Sang 2015;108:410-6. [Crossref] [PubMed]
- Huang CH, Reid ME, Blumenfeld OO. Exon skipping caused by DNA recombination that introduces a defective donor splice site into the human glycophorin A gene. J Biol Chem 1993;268:4945-52. [Crossref] [PubMed]
- Storry JR, Olsson ML. Genetic basis of blood group diversity. Br J Haematol 2004;126:759-71. [Crossref] [PubMed]
- Huang CH, Reid M, Daniels G, et al. Alteration of splice site selection by an exon mutation in the human glycophorin A gene. J Biol Chem 1993;268:25902-8. [Crossref] [PubMed]
- Storry JR, Reid ME, Fetics S, et al. Mutations in GYPB exon 5 drive the S–s–U+(var) phenotype in persons of African descent: implications for transfusion. Transfusion 2003;43:1738-47. [Crossref] [PubMed]
- Gassner C, Denomme GA, Portmann C, et al. Two Prevalent ∼100-kb GYPB deletions causative of the GPB-deficient blood group MNS phenotype S-s-U- in Black Africans. Transfus Med Hemother 2020;47:326-36. [Crossref] [PubMed]
- Lane WJ, Gleadall NS, Aeschlimann J, et al. Multiple GYPB gene deletions associated with the U– phenotype in those of African ancestry. Transfusion 2020;60:1294-307. [Crossref] [PubMed]
- Leffler EM, Band G, Busby GBJ, et al. Resistance to malaria through structural variation of red blood cell invasion receptors. Science 2017;356:eaam6393. [Crossref] [PubMed]
- Moores P. Four examples of the S–s–U–phenotype in an Indian family. Vox Sanguinis 1972;23:452-4. [Crossref] [PubMed]
- Kulkarni S, Jadhav S, Vasantha K, et al. Identification of rare S–s–U– blood group phenotype in RHD negative woman: first report from India. Vox Sang 2020;115:3-387. [Abstract P-497].
- Sondag-Thull D, Girard M, Blanchard D, et al. S-s-U-phenotype in a Caucasian family. Exp Clin Immunogenet 1986;3:181-6. [PubMed]
- Stern DA, Hawksworth DN, Watt JM, et al. A new low-frequency red cell antigen, 'SARAH'. Vox Sang 1994;67:64-7. [PubMed]
- McBean RS, Hyland CA, Hendry JL, et al. SARA: a "new" low-frequency MNS antigen (MNS47) provides further evidence of the extreme diversity of the MNS blood group system. Transfusion 2015;55:1451-6. [Crossref] [PubMed]
- Towns D, Hannon J, Hendry J, et al. Hemolytic disease of the fetus and newborn caused by an antibody to a low-prevalence antigen, anti-SARA. Transfusion 2011;51:1977-9. [Crossref] [PubMed]
- Venkataraman R, Yusuf K. Intravenous immunoglobulin in the management of a rare cause of hemolytic disease of the newborn: Anti-SARA antibodies. J Neonatal Perinatal Med 2017;10:329-32. [Crossref] [PubMed]
- Green C, Poole J, Ford D, et al. A postulated glycophorin B-A-B hybrid demonstrating heterogeneity of anti-Hop and anti-Nob sera. Transfus Med 1992;2:67. [Abstract P64].
- Lopez GH, Wei L, Ji Y, et al. GYP*Kip, a novel GYP(B-A-B) hybrid allele, encoding the MNS48 (KIPP) antigen. Transfusion 2016;56:539-41. [Crossref] [PubMed]
- Uchikawa M, Ogasawara K, Suzuki Y, et al. A new GP(B-A-B) hybrid molecule (GP.Yak) with Miltenberger Phenotype. Vox Sanguinis 2012;103:214. [Abstract P-464].
- Flower RL, Wei L, Ji YL, et al. GP.Kipp and GP.Yak BAB hybrid glycophorins: No difference in sequence or serology. Vox Sang 2013;105:10. [Abstract 2A-S02-02].
- Kaito S, Wang Z, Tanaka C, et al. Miltenberger related glycophorin phenotypes including GP.Hil (Mi.V) and antibodies to Hil and MINY antigens among Japanese donors. Vox Sang 2019;114:5-135. [Abstract P-88].
- Oda A, Suzuki Y, Isa K, et al. GP.Mot, a novel glycophorin hybrid molecule with miltenberger phenotype in Japanese blood donor. Vox Sang 2018;113:5-347. [Abstract P-548].
- Oda A, Suzuki Y, Isa K, et al. GP.MOT: A novel glycophorin variant identified in a Japanese blood donor. Transfusion 2021;61:2825-9. [Crossref] [PubMed]
- Lopez GH, Wilson B, Liew YW, et al. An alloantibody in a homozygous GYP*Mur individual defines JENU (MNS49), a new high-frequency antigen on glycophorin B. Transfusion 2017;57:716-7. [Crossref] [PubMed]
- Poole J, Warke N, King MJ, et al. An antibody to a novel glycophorin B epitope made in a GP.Mur/GP.Mur (MiIII/MiIII) individual. Transfus Med 2005;15:30-31. [Abstract SI46].
- Ito S, Kaito S, Miyazaki T, et al. A new antigen SUMI carried on glycophorin A encoded by the GYPA*M with c.91A>C (p.Thr31Pro) belongs to the MNS blood group system. Transfusion 2020;60:1287-93. [Crossref] [PubMed]
- Reid ME, Poole J, Green C, et al. MINY: A novel MNS-related blood group antigen. Vox Sanguinis 1992;63:129-32. [Crossref] [PubMed]
- Reid ME, Moore BPL, Poole J, et al. TSEN: A novel MNS-related blood group antigen. Vox Sanguinis 1992;63:122-8. [Crossref] [PubMed]
- Tippett P, Reid ME, Poole J, et al. The Miltenberger subsystem: is it obsolescent? Transfus Med Rev 1992;6:170-82. [Crossref] [PubMed]
- Chandanayingyong D, Pejrachandra S, Poole J. Three antibodies of the MNSs system and their association with the Miltenberger Complex of antigens. I. Anek serum. Vox Sang 1977;32:272-3. [Crossref] [PubMed]
- Reid ME, Lisowska E, Blanchard D. Section 3: Epitope determination of monoclonal antibodies to glycophorin A and glycophorin B. Coordinator's report. Antibodies to antigens located on glycophorins and band 3. Transfus Clin Biol 2002;9:63-72. [Crossref] [PubMed]
- Chen V, Halverson G, Wasniowska K, et al. Direct evidence for the existence of Miltenberger antigen. Vox Sang 2001;80:230-3. [Crossref] [PubMed]
- Fraser NS, Roots NM, Lopez GH, et al. Comparison of two monoclonal antibodies for the detection of Mia (MNS7) in MNS hybrid glycophorins. Pathology 2018;50 Suppl 1:abstr S104.
- Lopez GH, Wilson B, Turner RM, et al. Frequency of Mia (MNS7) and classification of Mia-positive hybrid glycophorins in an Australian blood donor population. Transfus Med Hemother 2020;47:279-86. [Crossref] [PubMed]
- Lin X, Rubio G, Patel J, et al. Hybrid glycophorin and red blood cell antigen genotyping in Asian American type O blood donors with Mia phenotype. Transfusion 2019;59:3767-75. [Crossref] [PubMed]
- Makroo RN, Bhatia A, Chowdhry M, et al. Frequency of Mi(a) antigen: a pilot study among blood donors. Indian J Med Res 2016;143:633-5. [Crossref] [PubMed]
- Mohn JF, Lambert RM, Rosamilia HG. Incidence of the blood group antigen Mia in the Caucasian and Negro populations of Western New York. Transfusion 1961;1:392-3. [Crossref] [PubMed]
- Lopez M, Kalvelage M, Billingsley K. Molecular detection of glycophorin A and B hybrid phenotypes for prediction of Mia antigen. Transfusion 2019;59:125A. [Abstract P-IM-26].
- Prathiba R, Lopez CG, Usin FM. The prevalence of GP Mur and anti-"Mia" in a tertiary hospital in Peninsula Malaysia. Malays J Pathol 2002;24:95-8. [PubMed]
- Chandanayingyong D, Pejrachandra S. Studies on the miltenberger complex frequency in Thailand and family studies. Vox Sanguinis 1975;28:152-5. [Crossref] [PubMed]
- Lin M, Broadberry RE. Immunohematology in Taiwan. Transfus Med Rev 1998;12:56-72. [Crossref] [PubMed]
- Wei L, Lopez GH, Zhang Y, et al. Genotyping analysis of MNS blood group GP(B-A-B) hybrid glycophorins in the Chinese Southern Han population using a high-resolution melting assay. Transfusion 2018;58:1763-71. [Crossref] [PubMed]
- Matson GA, Sutton HE, Swanson J, et al. Distribution of hereditary blood groups among Indians in South America. I. In Ecuador. Am J Phys Anthropol 1966;24:51-69. [Crossref] [PubMed]
- Matson GA, Sutton HE, Swanson J, et al. Distribution of hereditary blood groups among Indians in South America. VII. In Argentina. Am J Phys Anthropol 1969;30:61-83. [Crossref] [PubMed]
- Matson GA, Swanson J. Distribution of hereditary blood antigens among Indians in Middle America: III. In Guatemala. Am J Phys Anthropol 1963;21:301-17. [Crossref] [PubMed]
- Shah A, Jariwala K, Gupte S, et al. Pattern of distribution of 35 red cell antigens in regular voluntary blood donors of South Gujarat, India. Transfus Apher Sci 2018;57:672-5. [Crossref] [PubMed]
- Giles CM, Chandanayingong D, Webb AJ. Three antibodies of the MNSs system and their association with the Miltenberger complex of antigens. III. Anek, Raddon and Lane antisera in relation to each other and the Miltenberger complex. Vox Sang 1977;32:277-9. [Crossref] [PubMed]
- Wei L, Shan ZG, Flower RL, et al. The distribution of MNS hybrid glycophorins with Mur antigen expression in Chinese donors including identification of a novel GYP.Bun allele. Vox Sang 2016;111:308-14. [Crossref] [PubMed]
- Storry JR, Poole J, Condon J, et al. Identification of a novel hybrid glycophorin gene encoding GP. Hop. Transfusion 2000;40:560-5. [Crossref] [PubMed]
- Wei L, Lopez GH, Ji Y, et al. Genotyping for glycophorin GYP(B-A-B) hybrid genes using a single nucleotide polymorphism-based algorithm by matrix-assisted laser desorption/ionisation, time-of-flight mass spectrometry. Mol Biotechnol 2016;58:665-71. [Crossref] [PubMed]
- Kerdkaewngam K, Ovataga P, Tubrod J, et al. The MNS variants in Thai blood donors. Vox Sang 2018;113:228. [Abstract P-475].
- Huang CH, Kikuchi M, McCreary J, et al. Gene conversion confined to a direct repeat of the acceptor splice site generates allelic diversity at human glycophorin (GYP) locus. J Biol Chem 1992;267:3336-42. [Crossref] [PubMed]
- Grimsley S, Jongruamklang P, Jones B, et al. Monoclonal anti-s, clone P3BER does not recognize the s antigen in the context of GP(B-A-B) hybrid proteins GP.Mur and GP.Bun or GP(B-A) hybrid, GP.Hil. Transfus Med 2019;29:21. [Abstract SIM8].
- Jongruamklang P, Grimsley S, Thornton N, et al. Characterization of GYP*Mur and novel GYP*Bun-like hybrids in Thai blood donors reveals a qualitatively altered s antigen. Vox Sang 2020;115:472-7. [Crossref] [PubMed]
- Cleghorn TE. A memorandum on the miltenberger blood groups. Vox Sanguinis 1966;11:219-22. [Crossref] [PubMed]
- Roots NM, Fraser NS, Lopez GH, et al. GP.Mur red blood cells express variant form of s antigen (MNS4). Pathology 2018;50:abstr s103.
- Lomas-Francis C, Louzon M, Vege S, et al. The S antigen encoded by GYPB*Mit is a partial antigen: first report of alloanti-S in a person with S+ Mit+ RBCs. Vox Sang 2016;111:68. [Abstract 5B-S35-02].
- Mo Z, Li H, Huang L, et al. Prevalence and specificity of RBC alloantibodies in the general hospitalised population in Guangxi. Transfus Med 2015;25:313-9. [Crossref] [PubMed]
- Jansuwan S, Tangvarasittichai O, Tangvarasittichai S. Alloimmunization to red cells and the association of alloantibodies formation with splenectomy among transfusion-dependent β-thalassemia major/HbE patients. Indian J Clin Biochem 2015;30:198-203. [Crossref] [PubMed]
- Nadarajan VS. The prevalence, immunogenicity, and evanescence of alloantibodies to MUT and Mur antigens of GP.Mur red blood cells in a Southeast Asian patient cohort. Transfusion 2018;58:1189-98. [Crossref] [PubMed]
- Nakasone M, Achkar R, Scuracchio P, et al. The prevalence of anti-Mia in Brazilian patients: is it necessary to include Mia-positive RBCs in antibody screening routine? Transfusion 2019;59:124A. [Abstract P-IM-25].
- Poole J, Daniels G. Blood group antibodies and their significance in transfusion medicine. Transfus Med Rev 2007;21:58-71. [Crossref] [PubMed]
- Bakhtary S, Gikas A, Glader B, et al. Anti-Mur as the most likely cause of mild hemolytic disease of the newborn. Transfusion 2016;56:1182-4. [Crossref] [PubMed]
- Oak J, Mallari R, de Asis M, et al. Occult hemolytic anemia due to anti-Mur in a patient receiving blood from a region with a prominent Asian donor population. Transfusion. 2017;57:178A. [Abstract CP298].
- Hawkins P. The highs and lows of working in an IRL (Case Study 2). 2019. Available online: http://www.haabb.org/fall2019preview.html
- Velliquette R, Hu Z, Aeschlimann J, et al. The first reported case of an acute hemolytic transfusion reaction due to the rare specificity anti-Hut (anti-MNS19). Vox Sang 2020;115:3-387. [Abstract P-451].
- Mallari RA, Chan A, Powers RJ, et al. Fetal inheritance of GP*Mur causing severe HDFN in an unrecognized case of maternal alloimmunization. Transfusion 2020;60:870-4. [Crossref] [PubMed]
- Peyrard T. Molecular tools for investigating immunohaematology problems. ISBT Science Series 2015;10:31-8. [Crossref]
- Palacajornsuk P, Nathalang O, Tantimavanich S, et al. Detection of MNS hybrid molecules in the Thai population using PCR-SSP technique. Transfus Med 2007;17:169-74. [Crossref] [PubMed]
- Montgomery J, Wittwer CT, Palais R, et al. Simultaneous mutation scanning and genotyping by high-resolution DNA melting analysis. Nat Protoc 2007;2:59-66. [Crossref] [PubMed]
- Meyer S, Vollmert C, Trost N, et al. MNSs genotyping by MALDI-TOF MS shows high concordance with serology, allows gene copy number testing and reveals new St(a) alleles. Br J Haematol 2016;174:624-36. [Crossref] [PubMed]
- McBean RS, Hyland CA, Flower RL. Blood group genotyping: the power and limitations of the Hemo ID Panel and MassARRAY platform. Immunohematology 2015;31:75-80. [Crossref] [PubMed]
- Veldhuisen B, van der Schoot CE, de Haas M. Multiplex ligation-dependent probe amplification (MLPA) assay for blood group genotyping, copy number quantification, and analysis of RH variants. Immunohematology 2015;31:58-61. [Crossref] [PubMed]
- Ji Y, Wen J, Veldhuisen B, et al. Validation of the multiplex ligation-dependent probe amplification assay and its application on the distribution study of the major alleles of 17 blood group systems in Chinese donors from Guangzhou. Transfusion 2017;57:423-32. [Crossref] [PubMed]
- Goldman M, Núria N, Castilho LM. An overview of the Progenika ID CORE XT: an automated genotyping platform based on a fluidic microarray system. Immunohematology 2015;31:62-8. [Crossref] [PubMed]
- ID Core XT. Package Insert. 2018. Available online: https://www.fda.gov/media/117051/download
- Lane WJ, Westhoff CM, Uy JM, et al. Comprehensive red blood cell and platelet antigen prediction from whole genome sequencing: proof of principle. Transfusion 2016;56:743-54. [Crossref] [PubMed]
- Schoeman EM, Lopez GH, McGowan EC, et al. Evaluation of targeted exome sequencing for 28 protein-based blood group systems, including the homologous gene systems, for blood group genotyping. Transfusion 2017;57:1078-88. [Crossref] [PubMed]
- Schoeman EM, Roulis EV, Liew YW, et al. Targeted exome sequencing defines novel and rare variants in complex blood group serology cases for a red blood cell reference laboratory setting. Transfusion 2018;58:284-93. [Crossref] [PubMed]
- Mahmoud M, Gobet N, Cruz-Dávalos DI, et al. Structural variant calling: the long and the short of it. Genome Biol 2019;20:246. [Crossref] [PubMed]
Cite this article as: Lopez GH, Hyland CA, Flower RL. Glycophorins and the MNS blood group system: a narrative review. Ann Blood 2021;6:39.


