Rapid detection of bacterial contaminants in platelet concentrates using the next generation BACT/ALERT® VIRTUO® Microbial Detection System: improved safety and operational efficiency
Background
The storage conditions of platelets at room temperature under continuous agitation in gas-permeable bags make them more likely to support bacterial growth that can lead to septic transfusion reactions (STRs) (1,2).
Sources of bacterial contamination include the donor, both asymptomatic bacteremia and transient/commensal skin flora; nonsterile collection equipment; or during the production process (3,4). Several mitigation strategies have been put in place over the past several decades to improve platelet safety, including decreasing shelf-life; measures to prevent contamination at the time of collection (donor health questionnaires, improved skin disinfection, and diversion of initial blood volume); culture-based detection methods; rapid secondary detection methods; and pathogen reduction technologies (2,3,5,6). This review will focus on the evolution of culture-based methods for testing platelet concentrates including automated instrumentation.
Performing conventional colony count culture testing of platelet concentrates was considered both time-consuming and labor-intensive. An experimental microbial contamination screening method using a noninvasive adhesive label containing a carbon dioxide (CO2)-sensitive color indicator provided encouraging results as an alternative to the conventional culture method (7). The sensitivity was later found to be insufficient for quality control of platelet concentrates and automated culture-based systems were explored and found to be both sensitive and rapid (8-10).
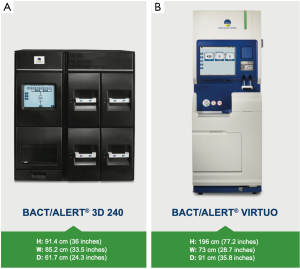
The BACT/ALERT® 3D (BTA 3D) microbial detection system was launched by Organon Teknika (acquired by bioMérieux in 2001) in 1997 and is the most prevalent automated culture system used to screen platelet concentrates (2,11,12) (Figure 1A). The BACT/ALERT® BPA (BPA, aerobic) and BACT/ALERT® BPN (BPN, anaerobic) culture bottles provide the necessary nutritional and environmental conditions for microorganisms that might be present in the test sample. The system utilizes a colorimetric sensor and reflected light to continuously monitor the presence and production of CO2 that is dissolved in the culture medium. If microorganisms are present in the test sample, CO2 is produced as the organisms metabolize the substrates in the culture medium. When growth of the microorganisms produce CO2, the color of the gas-permeable sensor installed in the bottom of each culture bottle changes to yellow (13).
Early evaluation studies using the BTA system indicated that ≥24 hours sampling time or increasing the sample volume (10 mL into each aerobic and anaerobic bottle) is necessary to provide confidence in bacterial detection (14-16). Early adoption of the BTA system for routine testing of platelet concentrates showed a reduction in outdating from 18.5% to 9.8% due to platelet shelf-life extension (17) and increased safety by preventing 1.8% contaminated leukocyte-reduced pooled platelet concentrates (18).
The National Blood Service evaluated the BTA system for testing leukocyte-reduced apheresis platelets (LRAP) and pooled buffy coat-derived platelet concentrates (19). They evaluated six microorganisms including Clostridium perfringens using both aerobic and anaerobic culture bottles at concentrations of between ~1 to 50 colony-forming units (CFU)/mL. Results showed rapid and substantial growth with all organisms with 100% detection within 48 hours and 98.1% detection within 24 hours. The C. perfringens strain tested was detected in all but one aerobic bottle, which posed the question if both aerobic and anaerobic culture is necessary or whether aerobic culture only is adequate.
Validation of the BTA system for screening platelet concentrates was performed with 15 microorganisms using the original and new BTA culture bottles (20,21). The new culture bottles utilize a colorimetric liquid emulsion sensor compared to the original solid-state colorimetric sensor disk and the new BPA bottle has an increased oxygen headspace that eliminates the need for venting. No difference in time to detection between the original and new culture bottles were found and with the exception of Cutibacterium (previously Propionibacterium) acnes, all organisms were detected between 9.2 to 20.4 hours when the starting concentration is ~10 to 100 CFU per mL. Cutibacterium acnes required a longer TTD and was detected in a mean time of 74.4 and 86.2 hours (100 and 10 CFU/mL, respectively). There was an overall 10% faster TTD with a starting concentration of 100 CFU/mL compared to 10 CFU/mL. The BTA system was validated again using nine microorganisms when the culture bottles were manufactured using plastic versus glass bottles (22). The data demonstrated that the new plastic bottles were superior or clinically comparable (within 0.1 h) to the glass BPA and BPN culture bottles.
A joint educational session by the American Society of Hematology and American Association of Blood Banks (AABB) provided a review of the risks, testing strategies, and regulatory approaches regarding bacterial contamination of blood components (5). This review was to help prepare for the new AABB Standard 5.1.5.1 included in its 22nd edition of Standards for Blood Banks and Transfusion Services that was adopted with final implementation date of March 1, 2004. The new standard stated, “The blood bank or transfusion service shall have methods to limit and detect bacterial contamination in all platelet components”. Automated culture-based methods were considered to be the most useful that was currently available and from a regulatory stand-point, the FDA encouraged manufacturers to submit applications for devices that will detect bacteria in transfusion products and include both internal and external (clinical trial) validation data. In December 2005, the FDA granted 510(k) clearance for BTA BPA (BK050037) and BTA BPN (BK050043) culture bottles to be used with the BTA 3D Systems for testing LRAP units, and both leukocyte-reduced single and a pool of up to six units of leukocyte-reduced whole blood platelet concentrates (LRWBPC).
From 2004–2008 several studies were published using the BTA 3D system for screening platelet concentrates using the typical culture method of 4 mL into an aerobic bottle only followed by quarantine for minimum of 12 hours before release for distribution as negative to date (23-27). These studies all concluded that the BTA 3D system could be used to identify and prevent transfusion of bacterially contaminated platelet units. Using only aerobic bottles to screen platelets was based on several factors including that platelets are stored under aerobic conditions; that clinically significant facultative anaerobic bacteria will grow under aerobic conditions; and lastly, most anaerobic bacteria encountered during platelet manufacture are not clinically significant (23).
The American Red Cross provided evidence of improved component safety by implementing inlet-line diversion, increased culture volume (4 to 8 mL), and improved skin disinfection (23,26-29). Their data collected from 2004 to 2008 showed that implementing inlet-line diversion decreased bacterial contamination during two-arm collections by more than 46% and by concurrently increasing the sample volume, from 4 to 8 mL the culture sensitivity increased 54%. In 2009, they found that using a single-step 2% chlorhexidine/70% isopropyl alcohol skin disinfection was more effective in preventing bacterial contamination of apheresis platelets compared to a two-step povidone-iodine method. In a similar study, Souza et al. (30) showed a trend of improved rate of detection (139 vs. 106 per million events), as well as a decrease in the mean-time of detection for true positives by 23% by increasing the sample volume from 4 to 8 mL in aerobic culture bottles. A meta-analysis of published North American data comparing bacterial contamination rates from 4 and 8 mL confirmed that higher sample volumes give higher true-positives and provide significant increase in detection rate and interdiction of contaminated units (31).
Canadian Blood Services (CBS), following a reported fatal septic shock associated with a platelet transfusion-transmitted Serratia marcescens, evaluated introduction of anaerobic culture bottles using 8–10 mL for each bottle and rapid secondary testing to determine the rate of detection failures (32). Testing was performed on outdated buffy-coat platelets that tested negative during initial screening. One true positive (Staphylococcus epidermidis) from a total of 4,002 platelets was found by both the BTA 3D and the immunoassay indicating that repeat screening should be considered to extend platelet storage. Data analysis from 6 years of testing at 12 CBS sites using the BTA 3D system and improved production protocols (increasing sample volume from 4–6 to 8–10 mL) did effectively reduce the risk of transfusion of bacterially contaminated platelet concentrates by increasing the initial positive and confirmed positive rates; however continued occurrence of false-negative results indicates a residual risk remains (33,34).
Screening for bacterial contamination using aerobic and anaerobic bottles was implemented in the Netherlands starting in 2001 (35) and in Australia in 2008 (36,37). Both Sanquin Blood Bank and Australian Red Cross Blood Service performed testing after 24 hours of collection using 7.5–10 mL into both aerobic and anaerobic bottles. Using 10 years of data, Sanquin Blood Bank reported a frequency of transfusion-transmitted bacterial infection of less than 1 per 2 years and the Australian Red Cross reported a 4.2-fold decline in reported STRs since introduction of bacterial screening using a two-bottle method.
A two-bottle system and secondary culture was introduced for bacterial screening of platelets at the Irish Blood Transfusion Service (IBTS) (38). The IBTS test using a large volume (10 mL) into both aerobic and anaerobic culture bottles on the day after manufacture and retest again using a large volume (7.5–10 mL) into aerobic and anaerobic culture bottles on day 4 of storage to allow extended storage of platelets. Using this culture method, they estimated that they could detect bacterial contamination at a concentration of 1 CFU/mL of platelet concentrate with greater than 99.5% sensitivity. A secondary bacterial culture method was initiated in October 2016 for all apheresis platelets received at The Johns Hopkins Hospital (JHH) (39). Primary culture was performed at the blood collection agency using 8 mL inoculation into an aerobic bottle 24 hours after donor collection. The platelet products are held in quarantine for 12 hours after sampling, after which they are released to JHH. Secondary culture is performed at the time of receipt (day 3 post-collection) and 5 mL samples of each platelet product is inoculated to a BPA culture bottle. The bottles are loaded into the BTA instrument and incubated for 3 days or until positive. During their 12-month study period, 93.5% platelet products were tested by secondary culture. A total of 8 positive cultures were reported (incidence of 1 in 2,881 platelet products). The authors report no STRs during the study period and the total cost was found to be more economically favorable than pathogen reduction and rapid secondary testing. In February 2018, the FDA granted 510(k) clearance for BTA BPA and BPN (BK170142) culture bottles as a secondary safety measure on the BACT/ALERT 3D.
Several enhanced primary culture methods were developed and implemented. A model based on the application of the Poisson distribution to detection of bacteria in platelet concentrates was introduced for sampling that uses a fixed proportion of the collection volume from single, double, and triple collections (40). This minimal proportional sample volume (MPSV) model samples at least 3.8% of mother bag volume and inoculates between 1 to 3 aerobic bottles (7–10 mL per bottle). The results showed improved sensitivity of primary culture and identified collections that could have escaped detection had only a single bottle with 8 to 10 mL volume been used (41). A large volume delayed sampling (LVDS) enhanced primary culture was implemented at the National Health Service Blood and Transplant (NHSBT) in 2011 (42). The NHSBT protocol samples at 36 to 48 hours after donation and 16 mL samples are taken with 8 mL inoculated to each of aerobic and anaerobic bottles, and a 6-hour holding period is used prior to distribution. From February 2011 to September 2015, a total of 1,239,029 platelet components were screened. The confirmed-positive rate was 0.03% and false-positive rate was 0.19%. They report four false-negative cultures (0.0003%), all with Staphylococcus aureus, three were visually detected before transfusion and one confirmed transmission resulted in patient morbidity. The screening protocol effectively reduced the number of clinically adverse transmissions by 90% during the reported time period compared to the 5 years before the introduction of bacterial screening of platelet components. Héma-Québec presented their LVDS protocol for screening platelets at the 2016 AABB Annual Meeting (43). They described an improved procedure that increases the volume from 10 to 20 mL; 10 mL into one aerobic and one anaerobic culture bottle. The delay before sampling was increased from 24 to 48 hours and a 12-hour holding period before labeling and distribution was introduced. Platelet shelf-life was allowed to 7 days. Their study showed a 71% reduction of outdates; an increase in positive culture rate (0.011% to 0.044%); and no STRs were recorded. Data collected from October 2015 to December 2017 showed a true positive rate of 0.035% (22/62,531). Five were transfused (Cutibacterium sp.) with no adverse reaction (44).
In November, 2017, The Blood Products Advisory Committee (BPAC) voted in favor of several culture strategies including enhanced primary (MPSV and LVDS) and secondary culture, as well as pathogen reduction and secondary rapid testing to improve platelet safety (45). These strategies were presented again to the FDA in July 2018 with an emphasis that having multiple strategies available will allow each organization to assess the economics and operational challenges to optimize patient safety and utilization (46). Each strategy has been shown to improve platelet safety and while contamination rates are low, residual risk remains (2,12,47,48). This is highlighted by the recent multi-state investigation involving sepsis caused by Acinetobacter calcoaceticus-baumannii complex and Staphylococcus saprophyticus that involved one platelet unit treated with pathogen-reduction technology, and two units that had tested negative with a rapid bacterial detection device after negative primary culture (49).
In September 2019 the FDA finalized its guidance to include both single-step and two-step strategies to extend the storage/dating period of platelets up to 5 days or up to 7 days depending on the strategy implemented in the laboratory (50). In March 2020, the FDA granted 510(k) clearance for BTA BPA and BPN (BK200472) (51) culture bottles for use on both BTA Detection Systems [BTA 3D and BACT/ALERT® VIRTUO® (BTA VIRTUO)] for quality control testing of LRAP units, and both single and pools of up to six units of LRWBPC, as a safety measure, to extend dating beyond day 5 and up to day 7 for LVDS of platelets no sooner than 48 hours after collection; or secondary culture no sooner than day 4 after platelet collection. In addition, the BTA Microbial Detection Systems can be used to extend dating to five days using LVDS of platelets no sooner than 36 hours after collection; or secondary culture no sooner than day 3 after platelet collection. The FDA updated the guidance in Dec 2020 to remove the LVDS footnote and to extend the initial implementation date of March 2021 to October 2021.
Extension of the shelf-life to 7 days has been estimated to have an improvement of 38% reduction in wastage (52). Deciding which strategy to implement is highly complex, because safety, operational efficiency, and cost are all considerations (47,53). Each strategy has different processing methods that result in different risks, efficacy, and viability that can affect platelet costs and availability. Recent literature focusing on the risks, benefits, and economics for the various strategies show wide variation in outcomes indicating that one size does not fit all (47,52,54-62). In a recent publication, CBS report that extension of platelet shelf-life to 7 days using a LVDS test method decreased septic transfusion events threefold, improved inventory, and reduced outdating by 10% (63). They calculated a net inferred cost benefit of implementing 7-day LVDS to be approximately $1.9 million.
The BTA VIRTUO Microbial Detection System is the next generation of BTA instrumentation (Figure 1B). As with previous generations of BACT/ALERT, the BTA VIRTUO has the capability to incubate, agitate, and continuously read BTA culture bottles. The underlying colorimetric technology used in previous generations of BTA is also used in the BTA VIRTUO. Enhancements of BTA VIRTUO (over previous BTA generations) incorporate new instrument architecture to improve temperature stability (minimizes occurrence of false-positives), workflow improvement via automation of processes that are currently performed manually, an improved user interface and an enhanced detection algorithm to shorten times to detection (TTD) of positive cultures. The enhanced algorithm uses multiple mathematical techniques to interpret changes in CO2. The mathematical methods used are able to rapidly and accurately detect a change in the shape of the curve that corresponds to the transition from the lag phase to log phase of microbial growth.
While numerous publications have been made on the BTA 3D systems for testing platelet concentrates, none have focused on the sensitivity, specificity, and improved efficiency of the BTA VIRTUO system. This review will include several internal and external studies using the BACT/ALERT microbial detection systems, specifically studies comparing the BTA 3D and BTA VIRTUO. The studies presented were performed to show analytical sensitivity, microbial recovery & reproducibility, repeatability, precision, and performance over platelet shelf-life, and improved laboratory efficiency with the BTA VIRTUO.
BACT/ALERT VIRTUO performance studies
For all of the studies presented, the operation of the BTA 3D and BTA VIRTUO Systems were performed according to the instructions provided in the instrument Operator’s Manuals and individual bottle Instructions for Use. All bottles remained in the BTA 3D or BTA VIRTUO instruments until signaled positive or negative by the instrument after 5–7 days of incubation, depending on the study. All positive bottles were sub-cultured under appropriate temperature and atmospheric conditions and at least one bottle per organism per lot, per instrument type had identification confirmed by either VITEK® MS or VITEK® 2 systems (bioMérieux).
The microorganisms used were selected based on published FDA reports, literature, and previous BTA 3D clinical studies and were prepared using either BIOBALL® SINGLESHOT (bioMérieux) or serially-diluted fresh stock cultures (Table 1).
Table 1
| Microorganism | Strain | Inoculum type | Study |
|---|---|---|---|
| Bacillus cereus | NCTC 7464 | BIOBALL®† | Sensitivity; buffy coats validation; recovery & reproducibility; repeatability |
| Clostridium perfringens (BPN only) | NCTC 8798 | BIOBALL | Sensitivity; buffy coats validation; recovery & reproducibility; repeatability |
| Enterobacter cloacae | ATCC® 35549™ | Stock Culture‡ | Sensitivity |
| ATCC® 29005™ | Stock Culture | Buffy coats validation; recovery & reproducibility | |
| Escherichia coli | NCTC 12241 | BIOBALL | Sensitivity; buffy coats validation; recovery & reproducibility; repeatability |
| Klebsiella pneumoniae | ATCC® 35657™ | Stock Culture | Sensitivity |
| ATCC® 8045™ | Stock Culture | Buffy coats validation; recovery & reproducibility | |
| Proteus mirabilis | ATCC® 7002™ | Stock Culture | Sensitivity |
| Pseudomonas aeruginosa (BPA only) | NCTC 12924 | BIOBALL | Sensitivity; repeatability |
| ATCC® 27853™ | Stock Culture | Buffy coats validation; recovery & reproducibility; repeatability | |
| Salmonella enterica subsp. enterica serovar Heidelberg | ATCC® 8326™ | Stock Culture | Buffy coats validation |
| Salmonella enterica subsp. enterica serovar Pomona | ATCC® 10729™ | Stock Culture | Sensitivity |
| Salmonella enterica subsp. enterica serotype Typhimurium | NCTC 12023 | BIOBALL | Sensitivity |
| Serratia liquefaciens | ATCC® 35551™ | Stock Culture | Buffy coats validation |
| Serratia marcescens | ATCC® 43862™ | Stock Culture | Sensitivity; recovery & reproducibility |
| Staphylococcus aureus | NCTC 10788 | BIOBALL | Sensitivity; buffy coats validation; recovery & reproducibility; repeatability |
| Staphylococcus epidermidis | NCTC 6513 | BIOBALL | Sensitivity; buffy coats validation; recovery & reproducibility |
| Streptococcus agalactiae | ATCC® 12927™ | Stock Culture | Buffy coats validation |
| Streptococcus pyogenes | NCTC 12696 | BIOBALL | Sensitivity; recovery & reproducibility; repeatability |
| Streptococcus sanguinis | ATCC® 10556™ | Stock Culture | Sensitivity; recovery & reproducibility |
†, BIOBALL SINGLESHOT has a batch mean of between 28–33 CFU. Multiple BIOBALL SINGLESHOT were dispensed directly into pooled platelet preparations such that the final inoculum to be inoculated was achieved. ‡, stock culture inoculum was prepared using serially diluted fresh culture. Microorganisms were serially diluted in appropriate medium with final dilution made in the platelet preparation to obtain a final organism concentration at the study target CFU per mL.
The data were used to summarize overall specificity (false positive rate) and sensitivity (false negative rate). Lastly, a time and motion study comparing the BTA 3D and BTA VIRTUO is presented to illustrate improved operational efficiency of the BTA VIRTUO system.
Analytical sensitivity: comparison of time to detection of the BACT/ALERT 3D and the BACT/ALERT VIRTUO
An internal inoculation study was performed with a panel of 13 microorganisms to establish equivalency between the time to detection of BTA 3D and BTA VIRTUO instruments when testing BPA/BPN culture bottles in the presence of LRAP (64). The LRAP in plasma units were obtained from the inventory of a platelet collection agency and were used within 5 days of collection. Aliquots of the pooled platelets were seeded with microorganisms with a target of 3–30 CFU/mL (Table 1).
In this study, 4 mL of the seeded LRAP aliquots were inoculated into 3 different lots of the appropriate BPA and BPN culture bottles and the bottles were tested on 1 BTA 3D and 3 BTA VIRTUO instruments. In addition, negative controls were tested at a volume of 10 mL of unseeded LRAP, per bottle type, per bottle lot across the 4 instruments.
The recovery and time to detection (TTD) for all of the tested microorganisms by bottle type and instrument are presented in Table 2. The platelets used in this study were confirmed free of bacterial contamination by the negative controls (data not shown). A total of 120 and 216 seeded culture bottles were tested on the BTA 3D and BTA VIRTUO, respectively. The detection and recovery for each instrument by bottle type was 100%. The overall TTD on the BTA VIRTUO was faster than or equivalent to the BTA 3D, with an average improvement of 2.6 hours for BPA bottles, and 2.7 hours for BPN bottles.
Table 2
| Microorganism† | Mean inoculum (CFU/mL) | Culture bottle | BACT/ALERT 3D, time to detection (hours)‡ | BACT/ALERT VIRTUO, time to detection (hours)‡ | |||
|---|---|---|---|---|---|---|---|
| Mean | Range | Mean | Range | ||||
| Bacillus cereus | 3 | BPA | 10.0 | 9.6–10.6 | 7.6 | 7.2–7.9 | |
| BPN | 12.9 | 12.2–13.9 | 11.0 | 9.5–12.7 | |||
| Clostridium perfringens | 2 | BPN | 9.4 | 8.9–10.3 | 7.3 | 6.6–7.8 | |
| Enterobacter cloacae | 1 | BPA | 23.6 | 22.1–25.2 | 22.2 | 18.3–26.6 | |
| BPN | 14.8 | 13.2–16.3 | 11.4 | 9.8–14.8 | |||
| Escherichia coli | 4 | BPA | 12.9 | 12.7–13.2 | 9.9 | 9.7–10.2 | |
| BPN | 11.8 | 11.5–12.0 | 9.3 | 8.9–10.3 | |||
| Klebsiella pneumoniae | 4 | BPA | 11.5 | 11.0–12.0 | 9.4 | 9.1–9.7 | |
| BPN | 12.0 | 11.8–12.2 | 9.6 | 9.4–10.1 | |||
| Proteus mirabilis | 20 | BPA | 14.5 | 13.0–15.8 | 11.5 | 10.3–13.5 | |
| BPN | 12.2 | 11.5–12.7 | 9.7 | 8.9–11.0 | |||
| Pseudomonas aeruginosa | 1 | BPA | 17.5 | 16.6–18.0 | 14.5 | 13.2–16.3 | |
| Salmonella enterica subsp. enterica serovar Pomona | 14 | BPA | 12.6 | 12.2–13.0 | 10.1 | 9.9–10.3 | |
| BPN | 11.5 | 11.3–11.8 | 9.3 | 8.9–9.6 | |||
| Salmonella enterica subsp. enterica serotype Typhimurium | 2 | BPA | 13.8 | 13.7–13.9 | 10.9 | 10.5–11.3 | |
| BPN | 12.3 | 12.0–12.5 | 9.8 | 9.0–10.6 | |||
| Serratia marcescens | 16 | BPA | 11.7 | 11.3–12.2 | 9.5 | 9.2–9.9 | |
| BPN | 12.1 | 11.8–12.5 | 10.0 | 9.7–10.3 | |||
| Staphylococcus aureus | 3 | BPA | 16.5 | 16.1–17.0 | 14.0 | 13.3–15.1 | |
| BPN | 17.8 | 17.3–18.5 | 14.4 | 13.9–14.9 | |||
| Staphylococcus epidermidis | 2 | BPA | 19.6 | 18.5–20.4 | 15.8 | 14.2–18.1 | |
| BPN | 22.2 | 21.6–22.6 | 18.5 | 17.4–20.0 | |||
| Streptococcus sanguinis | 25 | BPA | 21.0 | 19.2–22.6 | 16.6 | 15.3–19.5 | |
| BPN | 24.9 | 21.6–30.0 | 19.5 | 16.9–21.6 | |||
†, a total of 40 (20 BPA and 20 BPN) negative controls were tested at a volume of 10 mL of unseeded LRAP across the 4 instruments (1 BACT/ALERT 3D and 3 BACT/ALERT VIRTUO) to serve as negative controls and as a sterility test of the platelet units. All negative controls were declared negative by the instruments and negative upon subculture. ‡, a total of 120 seeded culture bottles (5 replicates per microorganism per bottle type) were tested on the BACT/ALERT 3D and a total of 216 seeded culture bottles (9 replicates per microorganism per bottle type) were tested on the BACT/ALERT VIRTUO. The detection and recovery for each instrument by bottle type was 100%.
The percent difference in TTD was determined by using the mean TTD for each microorganism per system combination (Table 3). Calculating the percent difference standardizes the data and allows comparison between faster and slower-growing microorganisms. The overall mean percent difference was calculated as well as a 95% confidence interval for the mean percent difference. The results in Table 3 show that the overall percent difference between the two systems is −19.2, which indicates improved performance of the BTA VIRTUO over the BTA 3D. The 95% confidence interval was calculated with an upper and lower limit range of −20.8 to −17.6, indicating the BTA VIRTUO demonstrated acceptable performance which was faster than the BTA 3D.
Table 3
| Microorganism | Comparison ratio, 100× (VIRTUO mean − BTA 3D mean)/BTA 3D mean | |
|---|---|---|
| BPA culture bottle | BPN culture bottle | |
| Bacillus cereus | −24.0 | −14.7 |
| Clostridium perfringens | Test not performed | −23.0 |
| Enterobacter cloacae | −5.9 | −21.2 |
| Escherichia coli | −23.3 | −20.0 |
| Klebsiella pneumoniae | −18.3 | −20.5 |
| Proteus mirabilis | −20.7 | −19.7 |
| Pseudomonas aeruginosa | −17.1 | Test not performed |
| Salmonella enterica subsp. enterica serovar Pomona | −19.8 | −19.5 |
| Salmonella enterica subsp. enterica serotype Typhimurium | −21.0 | −17.4 |
| Serratia marcescens | −18.8 | −19.1 |
| Staphylococcus aureus | −15.2 | −16.7 |
| Staphylococcus epidermidis | −19.4 | −21.7 |
| Streptococcus sanguinis | −21.0 | −23.2 |
| OVERALL percent difference (range using 95% confidence interval) | −19.2 (−20.8 to −17.6) | |
External studies: recovery & reproducibility of the BACT/ALERT 3D and the BACT/ALERT VIRTUO
Three external studies are presented. The first external study was a seeded buffy coat-derived platelet validation performed with a panel of microorganisms in BPA and BPN culture bottles tested in the BTA 3D and BTA VIRTUO instruments (64,65). Platelet preparations included pooled LRWB derived buffy coat platelets (buffy coats) in plasma, and pooled buffy coats in plasma plus platelet additive solution (PAS). Aliquots of platelet preparations were seeded with low levels of each microorganism at a target of 3–10 CFU/mL (Table 1), and inoculated into BPA/BPN culture bottles. A total of 10 replicates of each species per bottle type per system, plus a minimum of 200 negative (platelets only, no organism) controls per bottle type per system was performed. A total of 400 culture bottles were seeded with 8 mL of each of the two buffy coat preparations with microorganisms and were loaded equally into each instrument and incubated until declared positive by the instruments or for up to 7 days. Seeded bottle data were used to evaluate the differences in the overall recovery rates between instruments. The negative control bottles from each instrument were tested to evaluate differences in the overall negative agreement rates (detection of false positives) between instruments and to serve as sterility controls for the platelet preparations.
The buffy coats validation result summaries are shown in Tables 4 and 5. Overall, both systems (BTA VIRTUO and BTA 3D) detected 400/400 (100%) of the seeded bottles with buffy coats regardless of bottle type or platelet preparation type. The negative agreement rate for buffy coats for BTA 3D was 99.5% (210/211), and for BTA VIRTUO was 100% (210/210). One BPN negative control bottle on the BTA 3D turned positive with buffy coats in PAS. There was no difference in negative agreement rates between systems (P>0.05). Overall, for 800 tests with buffy coats, the mean TTD for microorganisms tested in BTA 3D was 13.6 hours (n=400) and in BTA VIRTUO 11.5 hours (n=400). BTA VIRTUO was faster than BTA 3D, overall, in detecting microorganisms with a difference (mean) of 2.1 hours (P<0.001).
Table 4
| Microorganism | Culture bottle | Inoculum (CFU/mL) | Number of positive cultures† | Mean time to detection (hours) | |||
|---|---|---|---|---|---|---|---|
| BTA 3D | VIRTUO | BTA 3D | VIRTUO | ||||
| Bacillus cereus | BPA | 4 | 10 | 10 | 10.1 | 8.1 | |
| BPN | 2 | 10 | 10 | 10.7 | 8.5 | ||
| Clostridium perfringens | BPN | 2 | 10 | 10 | 11.4 | 8.9 | |
| Enterobacter cloacae | BPA | 23 | 10 | 10 | 14.8 | 12.6 | |
| BPN | 23 | 10 | 10 | 11.4 | 8.9 | ||
| Escherichia coli | BPA | 4 | 10 | 10 | 12.7 | 10.3 | |
| BPN | 3 | 10 | 10 | 11.6 | 9.9 | ||
| Klebsiella pneumoniae | BPA | 10 | 10 | 10 | 15.3 | 13.7 | |
| BPN | 8 | 10 | 10 | 12.9 | 11.1 | ||
| Pseudomonas aeruginosa | BPA | 14 | 10 | 10 | 16.7 | 13.8 | |
| Salmonella enterica subsp. enterica serovar Heidelberg | BPA | 14 | 10 | 10 | 15.6 | 12.6 | |
| BPN | 13 | 10 | 10 | 10.9 | 8.7 | ||
| Serratia liquefaciens | BPA | 1 | 10 | 10 | 16.1 | 14.0 | |
| BPN | 2 | 10 | 10 | 14.6 | 13.4 | ||
| Staphylococcus aureus | BPA | 2 | 10 | 10 | 18.0 | 15.2 | |
| BPN | 4 | 10 | 10 | 15.7 | 13.9 | ||
| Staphylococcus epidermidis | BPA | 3 | 10 | 10 | 17.5 | 15.3 | |
| BPN | 3 | 10 | 10 | 18.9 | 16.1 | ||
| Streptococcus agalactiae | BPA | 20 | 10 | 10 | 10.9 | 8.3 | |
| BPN | 21 | 10 | 10 | 10.2 | 7.9 | ||
| None | BPA | <1‡ | 0 | 0 | – | – | |
| BPN | <1 | 0 | 0 | – | – | ||
†, 10 replicates of each species per bottle type per system was tested. A minimum of 200 bottles in each instrument (platelets only, no microorganism) were tested to evaluate false-positive rates. ‡, below the level of detection.
Table 5
| Microorganism | Culture bottle | Inoculum (CFU/mL) | Number of positive cultures† | Mean time to detection (hours) | |||
|---|---|---|---|---|---|---|---|
| BTA 3D | VIRTUO | BTA 3D | VIRTUO | ||||
| Bacillus cereus | BPA | 3 | 10 | 10 | 9.8 | 8.0 | |
| BPN | 2 | 10 | 10 | 11.7 | 9.9 | ||
| Clostridium perfringens | BPN | 3 | 10 | 10 | 10.3 | 8.4 | |
| Enterobacter cloacae | BPA | 21 | 10 | 10 | 12.3 | 9.8 | |
| BPN | 16 | 10 | 10 | 11.4 | 9.0 | ||
| Escherichia coli | BPA | 2 | 10 | 10 | 12.1 | 10.2 | |
| BPN | 3 | 10 | 10 | 11.1 | 9.6 | ||
| Klebsiella pneumoniae | BPA | 9 | 10 | 10 | 13.5 | 11.6 | |
| BPN | 9 | 10 | 10 | 13.0 | 11.1 | ||
| Pseudomonas aeruginosa | BPA | 18 | 10 | 10 | 16.0 | 13.7 | |
| Salmonella enterica subsp. enterica serovar Heidelberg | BPA | 24 | 10 | 10 | 12.0 | 9.8 | |
| BPN | 19 | 10 | 10 | 10.9 | 9.0 | ||
| Serratia liquefaciens | BPA | 1 | 10 | 10 | 16.3 | 14.2 | |
| BPN | 2 | 10 | 10 | 16.8 | 15.1 | ||
| Staphylococcus aureus | BPA | 4 | 10 | 10 | 17.3 | 14.8 | |
| BPN | 3 | 10 | 10 | 16.2 | 14.2 | ||
| Staphylococcus epidermidis | BPA | 4 | 10 | 10 | 17.9 | 16.0 | |
| BPN | 3 | 10 | 10 | 18.9 | 18.8 | ||
| Streptococcus agalactiae | BPA | 19 | 10 | 10 | 10.4 | 8.6 | |
| BPN | 19 | 10 | 10 | 10.2 | 8.3 | ||
| None | BPA | <1‡ | 0 | 0 | – | – | |
| BPN | <1 | 1 | 0 | – | – | ||
†, 10 replicates of each species per bottle type per system was tested. A minimum of 200 bottles in each instrument (platelets only, no microorganism) were tested to evaluate false-positive rates. ‡, below the level of detection.
A second multicenter external reproducibility study was performed using the BTA 3D and BTA VIRTUO to determine the ability of the BPA and BPN culture bottles to detect the presence of microorganisms in LRAP in plasma only (66). At each site, aliquots of the platelets were seeded with microorganisms at a target inoculum of 5–10 CFU/mL (Table 1). Due to the high volume of LRAP units required for the study, 4 mL of the seeded LRAP aliquots were inoculated into the appropriate BPA and BPN bottles and the bottles were tested in the BTA 3D and BTA VIRTUO systems. A total of 10 repetitions per system were performed for each microorganism per bottle type. The bottles were loaded into each instrument and incubated until declared positive by the instruments or up to 7 days. The overall recovery and detection rates were compared. Additionally, 419 bottles were tested in both instruments (208 in BTA 3D and 211 in BTA VIRTUO), using 10 mL per bottle of LRAPs only (without organism) to act as negative/sterility controls for the platelet preparations and to evaluate false positive rates between instruments.
A total of 800 BTA culture bottles (100 per instrument and bottle type at each site) were inoculated from seeded LRAPs. The recovery and TTD for test microorganisms by bottle type and instrument are summarized in Table 6. The BTA VIRTUO detected 398/400 (99.5%) and BTA 3D detected 400/400 (100%) of the seeded bottles. The 2 seeded bottles not detected as positive on the BTA VIRTUO were negative on subculture and gram stain. Repeat testing duplicates were status positive and were considered in agreement with the expected result. For both the BPA and BPN culture bottles, the authors determined no difference in recovery rates between systems by bottle type or overall (P>0.05). The BTA VIRTUO was negative for 211/211 (100%) and BTA 3D was negative for 207/208 (99.5%) of the total unseeded BPA/BPN culture bottles. One BPN culture bottle flagged positive on the BTA 3D system that was negative for bacterial growth on subculture. The authors determined no difference in negative agreement rates between systems by bottle type or overall (P>0.05). The TTD was found to be faster with the BTA VIRTUO system for all bacterial species tested in both BPA and BPN bottles. The overall improved TTD for BTA VIRTUO was found to be 2.8 hours with BPA and 3.1 hours with BPN (P<0.001).
Table 6
| Microorganism | Culture bottle† | Inoculum ranges (CFU/mL) | Number of positive cultures | Mean time to detection (hours) | |||
|---|---|---|---|---|---|---|---|
| BTA 3D | VIRTUO | BTA 3D | VIRTUO | ||||
| Bacillus cereus | BPA | 1–7 | 20 | 19 | 10.3 | 7.7§ | |
| BPN | 1–8 | 20 | 20 | 11.8 | 9.4 | ||
| Clostridium perfringens | BPN | 1–7 | 20 | 20 | 11.1 | 8.0 | |
| Enterobacter cloacae | BPA | 3–16 | 20 | 20 | 12.7 | 10.0 | |
| BPN | 4–20 | 20 | 20 | 11.6 | 9.0 | ||
| Escherichia coli | BPA | 1–8 | 20 | 20 | 12.6 | 9.6 | |
| BPN | 1–7 | 20 | 20 | 11.9 | 9.2 | ||
| Klebsiella pneumoniae | BPA | 2–17 | 20 | 20 | 14.5 | 11.7 | |
| BPN | 3–19 | 20 | 20 | 13.8 | 11.2 | ||
| Pseudomonas aeruginosa | BPA | 2–20 | 20 | 20 | 16.7 | 13.3 | |
| Serratia marcescens | BPA | 2–14 | 20 | 20 | 12.4 | 10.2 | |
| BPN | 1–12 | 20 | 20 | 12.5 | 10.2 | ||
| Staphylococcus aureus | BPA | 1–10 | 20 | 20 | 17.1 | 14.5 | |
| BPN | 2–6 | 20 | 20 | 15.7 | 12.5 | ||
| Staphylococcus epidermidis | BPA | 1–7 | 20 | 20 | 20.4 | 18.1 | |
| BPN | 1–8 | 20 | 20 | 17.7 | 14.6 | ||
| Streptococcus pyogenes | BPA | 1–7 | 20 | 20 | 15.0 | 12.2 | |
| BPN | 2–9 | 20 | 20 | 13.9 | 11.2 | ||
| Streptococcus sanguinis | BPA | 1–16 | 20 | 20 | 21.8 | 17.4 | |
| BPN | 1–17 | 20 | 19 | 27.0 | 20.7§ | ||
| Positive | BPA | – | 200 | 199 | – | – | |
| BPN | – | 200 | 199 | – | – | ||
| Total % recovery (95% confidence interval) | BPA | – | 100% (98.2–100%) | 99.5% (97.2–99.9%) | – | – | |
| BPN | – | 100% (98.2–100%) | 99.5% (97.2–99.9%) | – | – | ||
| No organism‡ (95% confidence interval) | BPA | <1 | 0/104 (0–3.5%) | 0/106 (0–3.4%) | – | – | |
| BPN | <1 | 1/104 (0.02–5.2%) | 0/105 (0–3.5%) | – | – | ||
†, ten replicates of each bottle type at each external testing site were inoculated with each microorganism at the inoculum level indicated. ‡, one BACT/ALERT BPA and BPN bottle for each system was inoculated with 10 mL of unseeded platelets to serve as negative controls each time seeded tested was performed. These negative controls served as a sterility test (<1 = below the level of detection) and to establish false positive rates. §, mean of 19 replicates.
Lastly, a reproducibility study was performed at three sites (one internal and two external), testing LRAP in plasma and a panel of 6 microorganisms at a target of 5 CFU/mL (Table 1). Each site performed the seeded study using the BTA VIRTUO instrument only, 2 BPA and BPN lot numbers, and 2 different operators performing the tests. Four mL of the seeded LRAP aliquots were inoculated into the appropriate BPA and BPN culture bottles and the bottles were loaded into each instrument and incubated until declared positive by the BTA VIRTUO instruments or up to 7 days. Negative (10 mL unseeded LRAP) controls were included to determine that no contamination or false positives were introduced by the platelet material.
Overall, there were 270 seeded and 54 unseeded bottles tested at the 3 sites. Table 7 shows the percent recovery for each tested microorganism as the overall percent recovery by site and for all sites combined. The mean TTD and corresponding range are shown for each microorganism for all sites, as well as the actual inoculum ranges. The BTA VIRTUO reproducibility study results show 100% recovery of all microorganisms for all sites combined with no false positive results recorded.
Table 7
| Microorganism† | Inoculum range (CFU/mL) | % recovery (number inoculated/number positive) | Time to detection (hours) | ||||
|---|---|---|---|---|---|---|---|
| Site 1 | Site 2 | Site 3 | Mean | Range | |||
| Bacillus cereus | 1–7 | 100% (18/18) | 100% (18/18) | 100% (18/18) | 8.5 | 7.2–13.3 | |
| Clostridium perfringens | 1–9 | 100% (9/9) | 100% (9/9) | 100% (9/9) | 7.9 | 6.6–9.8 | |
| Escherichia coli | 1–7 | 100% (18/18) | 100% (18/18) | 100% (18/18) | 9.3 | 7.8–10.3 | |
| Pseudomonas aeruginosa | 1–20 | 100% (9/9) | 100% (9/9) | 100% (9/9) | 13.2 | 12.5–14.2 | |
| Staphylococcus aureus | 1–7 | 100% (18/18) | 100% (18/18) | 100% (18/18) | 13.2 | 9.6–15.9 | |
| Streptococcus pyogenes | 1–7 | 100% (18/18) | 100% (18/18) | 100% (18/18) | 11.7 | 9.8–16.9 | |
†, three negative control bottles (1 BPA and 2 BPN or 2 BPA and 1 BPN depending upon the run) were inoculated with 10 mL of unseeded platelets by each operator each day to serve as negative controls and incubated along with the seeded bottles. A total of 54 culture bottles (27 BPA and 27 BPN) were inoculated and overall negative agreement rate was 100% (54/54).
Within laboratory precision (repeatability) of BACT/ALERT BPA and BPN culture bottles on BACT/ALERT VIRTUO Microbial Detection System
An internal study was performed during instrument validation to establish evidence of repeatability of growth performance of the BPA and BPN culture bottles tested with 4 mL of LRAP over 10 days (not consecutive), with two teams of operators on the BTA VIRTUO. Bottles were also tested on the BTA 3D, for reference. The LRAP in plasma units were obtained from the inventory of a platelet collection agency and were used within 5 days of collection. Aliquots of the platelets were seeded with six microorganisms at a target inoculum of 5 CFU/mL (Table 1).
Testing was performed using 3 lots of BPA and BPN culture bottles. For a given bottle type, 4 bottles of each lot were inoculated with 4 mL of seeded LRAP and the bottles were tested on 1 BTA 3D and 3 BTA VIRTUO instruments. In addition, a total of 250 negative controls were tested at a volume of up to 10 mL of unseeded LRAP, per bottle type, per bottle lot across the 4 instruments. The volume was variable due to platelet availability from the supplier.
The recovery and TTD for all of the tested microorganisms by bottle type and instrument are presented in Table 8. A total of 900 seeded culture bottles were tested on the BTA VIRTUO. Of these, 899 (99.9%) were declared positive. A total of 300 seeded culture bottles were tested on the BTA 3D. Of these, 299 (99.7%) were declared positive. The rate of detection and recovery between the BTA VIRTUO and BTA 3D was equivalent. Overall the TTD on the BTA VIRTUO was less than or equivalent to the BTA 3D with an average improvement in TTD of 2.8 hours when tested with BPA bottles and 2.9 hours when tested with BPN bottles. The BTA VIRTUO achieved an overall improved TTD of 2.8 hours, when compared to the BTA 3D results in the same study.
Table 8
| Microorganism† | Mean inoculum (CFU/mL) | Culture bottle | BACT/ALERT 3D | BACT/ALERT VIRTUO | |||||
|---|---|---|---|---|---|---|---|---|---|
| % recovery | Time to detection (hours) | % recovery | Time to detection (hours) | ||||||
| Mean | Range | Mean | Range | ||||||
| Bacillus cereus | 4 | BPA | 100 | 10.1 | 9.6–10.8 | 100 | 7.6 | 7.0–8.3 | |
| BPN | 100 | 14.1 | 11.3–16.3 | 100 | 10.8 | 7.7–13.4 | |||
| Clostridium perfringens | 4 | BPN | 96.7 | 13.8 | 8.6–102.2‡ | 100 | 10.0 | 6.5–62.4‡ | |
| Escherichia coli | 3 | BPA | 100 | 12.8 | 12.0–13.7 | 100 | 10.0 | 9.2–11.2 | |
| BPN | 100 | 11.8 | 11.3–12.7 | 100 | 9.3 | 8.6–10.5 | |||
| Pseudomonas aeruginosa | 2§ | BPA | 100 | 17.4 | 15.6–18.2 | 98.9 | 14.1 | 12.6–16.4 | |
| Staphylococcus aureus | 3 | BPA | 100 | 16.5 | 15.6–18.0 | 100 | 13.8 | 12.4–15.1 | |
| BPN | 100 | 17.6 | 16.8–18.5 | 100 | 14.4 | 13.1–16.5 | |||
| Streptococcus pyogenes | 3 | BPA | 100 | 15.8 | 14.2–17.0 | 100 | 13.0 | 11.1–15.5 | |
| BPN | 100 | 12.8 | 12.0–13.7 | 100 | 10.3 | 9.6–11.0 | |||
†, a total of 250 negative control bottles were tested with volumes from 4–10 mL of unseeded LRAP [62 on BACT/ALERT 3D (31 BPA and 31 BPN) and 188 on BACT/ALERT VIRTUO (96 BPA and 92 BPN)] to assess the risk of false positives. All negative controls were declared negative by the instruments after 7 days and upon subculture. No false positive bottles were observed. ‡, C. perfringens demonstrated sporadic, prolonged time-to-detection in some bottles on each detection system. Bottles were subcultured and identified as C. perfringens.§, for two tested BPA and BPN bottle lots all but one of the 10 days the plate counts for P. aeruginosa (BIOBALL) were 0, but out of the 80 bottles tested, only one bottle was declared negative and was subculture negative. All other bottles were declared positive, subcultured and demonstrated morphology consistent with P. aeruginosa. For the 3rd BPA and BPN bottle lot, a stock culture suspension of P. aeruginosa was prepared and used.
Performance of the BACT/ALERT 3D and BACT/ALERT VIRTUO systems for use as a secondary or safety measure test to extend the shelf life of platelet preparations
Internal and Clinical studies have demonstrated the BACT/ALERT VIRTUO to be equivalent to the BACT/ALERT 3D for detecting bacterial contamination in LRAP (Tables 2,6-8). To establish the equivalency of the BTA VIRTUO to the BTA 3D as a safety measure for secondary testing of previously tested platelet products, a statistical analysis was performed to show that the age of platelets does not affect the TTD of organisms representing transfusion relevant contaminants (67). This analysis was performed to show that the BTA VIRTUO is an effective secondary or safety measure for retesting of previously tested platelet products for the purpose of extending the outdating. The experimental set-up and data used in this analysis is outlined under the within laboratory precision (repeatability) study.
An analysis of variance based on a multi-sample median test (Brown-Mood test) was performed to determine whether the TTD was significantly different depending on the age of the platelets. A chi-square statistical analysis was used to determine whether platelet age has a significant effect on TTD. P values greater than 0.05 indicate that there is no statistically significant evidence indicating that platelet age effects TTD. A separate median test was completed for each organism/bottle combination. All P values for both the BTA 3D and BTA VIRTUO were greater than 0.05 (Table 9), providing confirmation that platelet age does not affect TTD.
Table 9
| Instrument | Microorganism† | Culture bottle | Platelet age (days) | N | Mean TTD (hours) | Median TTD (hours) | Median test χ2 Exact P value |
|---|---|---|---|---|---|---|---|
| BACT/ALERT 3D | Bacillus cereus | BPA | 3 | 4 | 10.28 | 10.10 | 0.2191 |
| 4 | 10 | 9.92 | 9.80 | ||||
| 5 | 6 | 10.05 | 10.10 | ||||
| BPN | 3 | 4 | 14.70 | 14.40 | 0.6317 | ||
| 4 | 10 | 15.06 | 15.10 | ||||
| 5 | 6 | 14.12 | 14.40 | ||||
| Clostridium perfringens | BPN | 3 | 3 | 13.67 | 10.30 | 0.5772 | |
| 4 | 10 | 19.83 | 10.70 | ||||
| 5 | 6 | 11.78 | 10.30 | ||||
| Escherichia coli | BPA | 3 | 4 | 12.98 | 13.10 | 0.4137 | |
| 4 | 10 | 12.86 | 12.70 | ||||
| 5 | 6 | 12.68 | 12.60 | ||||
| BPN | 3 | 4 | 11.70 | 11.65 | 0.4622 | ||
| 4 | 10 | 11.59 | 11.50 | ||||
| 5 | 6 | 11.77 | 11.80 | ||||
| Pseudomonas aeruginosa | BPA | 3 | 4 | 17.33 | 17.25 | 0.8363 | |
| 4 | 10 | 17.34 | 17.40 | ||||
| 5 | 6 | 17.22 | 17.00 | ||||
| Staphylococcus aureus | BPA | 3 | 4 | 16.25 | 16.30 | 0.8687 | |
| 4 | 10 | 16.61 | 16.45 | ||||
| 5 | 6 | 16.72 | 16.70 | ||||
| BPA | 3 | 4 | 17.50 | 17.40 | 0.9378 | ||
| 4 | 10 | 17.68 | 17.60 | ||||
| 5 | 6 | 17.50 | 17.40 | ||||
| Streptococcus pyogenes | BPA | 3 | 4 | 16.00 | 15.95 | 0.6006 | |
| 4 | 10 | 16.20 | 16.10 | ||||
| 5 | 6 | 16.35 | 16.45 | ||||
| BPN | 3 | 4 | 12.80 | 12.85 | 0.7646 | ||
| 4 | 10 | 12.99 | 14.00 | ||||
| 5 | 6 | 12.97 | 13.00 | ||||
| BACT/ALERT VIRTUO | Bacillus cereus | BPA | 3 | 12 | 7.50 | 7.50 | 0.4592 |
| 4 | 30 | 7.59 | 7.55 | ||||
| 5 | 18 | 7.50 | 7.50 | ||||
| BPN | 3 | 12 | 11.54 | 11.55 | 0.0928 | ||
| 4 | 30 | 11.30 | 11.50 | ||||
| 5 | 18 | 10.56 | 10.95 | ||||
| Clostridium perfringens | BPN | 3 | 12 | 13.42 | 8.10 | 0.6948 | |
| 4 | 30 | 11.11 | 8.05 | ||||
| 5 | 18 | 10.58 | 7.95 | ||||
| Escherichia coli | BPA | 3 | 12 | 9.81 | 9.80 | 0.7230 | |
| 4 | 30 | 9.84 | 9.85 | ||||
| 5 | 18 | 9.79 | 9.75 | ||||
| BPN | 3 | 12 | 9.21 | 9.10 | 0.2486 | ||
| 4 | 30 | 9.19 | 9.15 | ||||
| 5 | 18 | 9.04 | 9.00 | ||||
| Pseudomonas aeruginosa | BPA | 3 | 12 | 13.97 | 14.05 | 0.2076 | |
| 4 | 29 | 13.98 | 13.80 | ||||
| 5 | 18 | 13.67 | 13.70 | ||||
| Staphylococcus aureus | BPA | 3 | 12 | 13.61 | 13.50 | 0.3623 | |
| 4 | 30 | 13.85 | 13.90 | ||||
| 5 | 18 | 13.79 | 13.95 | ||||
| BPA | 3 | 12 | 14.22 | 14.20 | 0.2226 | ||
| 4 | 30 | 14.31 | 14.30 | ||||
| 5 | 18 | 14.04 | 14.00 | ||||
| Streptococcus pyogenes | BPA | 3 | 12 | 13.48 | 13.40 | 0.8416 | |
| 4 | 30 | 13.70 | 13.80 | ||||
| 5 | 18 | 13.66 | 13.65 | ||||
| BPN | 3 | 12 | 10.38 | 10.45 | 0.4334 | ||
| 4 | 30 | 10.30 | 10.35 | ||||
| 5 | 18 | 10.17 | 10.10 |
†, a total of 250 negative control bottles were tested with volumes from 4–10 mL of unseeded LRAP. All negative controls were declared negative by the instruments after 7 days and upon subculture. No false positive bottles were observed.
Specificity and sensitivity
The BACT/ALERT VIRTUO System overall specificity (false positive rate) and sensitivity (false negative rate) were determined from internal and external studies conducted during performance validation utilizing LRAP and reported in Tables 2,6-8.
During the performance validation testing for LRAP, negative controls containing 4–10 mL of platelet concentrate were tested. No instrument false positives were observed during the performance validation. Results for the negative controls are summarized in Table 10. A total of 485 negative controls were performed and there were no false positive results recorded.
Table 10
The sensitivity of the BTA VIRTUO instrument was determined during performance validation testing for LRAP. All negative control bottles and any seeded bottles that were determined negative by the instrument were confirmed to be true negatives through subculture to plated media. There were no false negative bottles observed from the 1,786 seeded culture bottles.
Time and motion study: BACT/ALERT 3D versus VIRTUO
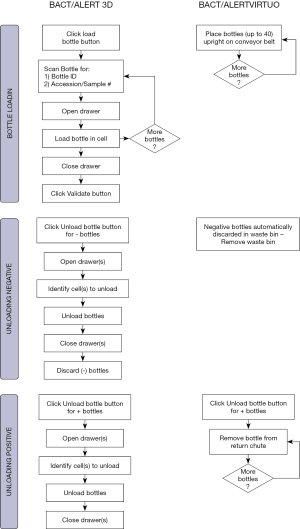
The BTA VIRTUO incorporates a number of improved features such as automated loading and unloading of culture bottles, and an enhanced algorithm that has been shown to reduce the time to detection (64,66,67). A time and motion study comparing the BTA VIRTUO and BTA 3D instruments was performed in order to evaluate the standard times of the different activities made by a laboratory technician when running a culture test with an automated culture system (68). The objective of the study was to determine if the new BTA VIRTUO platform could optimize workflow by providing significant time savings.
The study was performed using a Certified Lean-Six Sigma Black Belt Consultant and scientists trained in the use of the BTA VIRTUO and BTA 3D systems and familiar with the normal workflow within a laboratory performing quality control testing on LRAP Units.
Time measurements were collected to determine the observed time for loading and unloading culture bottles. Loading studies used a total of 40 bottles while unloading studies used 35 negative bottles and 7 positive bottles. The number of steps and time savings were evaluated to identify potential savings including hands-on-time, workload, and workers. MINITAB version 18 statistical software was used to determine significance.
The BTA 3D and BTA VIRTUO instruments are shown in Figure 1. A full bank (4 units) BTA VIRTUO configuration holds 272 more bottles compared to a BTA 3D 1440 system (6 Incubation Modules). The dimensions of a full BTA VIRTUO bank are: H: 196 cm (77.2 inches) × W: 292 cm (114.8 inches) × D: 91 cm (35.8 inches). In comparison, a BTA 3D with 6 Incubation Modules and one Controller Module optimized by utilizing stacking carts to configure Incubation Modules one on top another has the following dimensions including required stacking carts: H: 199.4 cm (78.5 inches) × W: 203.2 cm (80 inches) × D: 63.5 cm (25 inches) and 114.3 cm (45 inches) with drawer open (required for loading and unloading bottles). Because BTA VIRTUO does not require opening and closing incubator drawers, the BTA VIRTUO design reduces the instrument footprint for normal user activity by 30%.
The BTA 3D system requires 17 steps and 18 seconds per bottle to perform culture testing while the BTA VIRTUO system requires only 3 steps and 3 seconds per bottle to perform the same testing (Figure 2, Tables 11,12). The mean of the BTA 3D loading time and time to unload positive bottles were found to be greater than those with BTA VIRTUO at the 0.05 level of significance. The 14 steps and 15 seconds reduction to handle bottles with the BTA VIRTUO system is due to the automated conveyor loading and direct waste removal of negative culture bottles.
Table 11
| Process in steps | Loading bottles | Negative bottle management | Positive bottle management | Grand total |
|---|---|---|---|---|
| BTA 3D number of steps | 6 | 6 | 5 | 17 |
| VIRTUO number of steps | 1 | 0† | 2 | 3 |
| Savings in steps | 5 | 6 | 3 | 14 |
| Savings in % | 83 | 100 | 60 | 82 |
†, auto unload. Waste bin capacity is 7 kg or 72 bottles.
Table 12
| Process in seconds | Loading time per bottle | Unloading negatives per bottle | Unloading positives per bottle | Total time |
|---|---|---|---|---|
| BTA 3D time consumed in s | 9 | 4 | 5 | 18 |
| VIRTUO time consumed in s | 1 | 0 | 2 | 3 |
| Savings in time (s) | 8 | 4 | 3 | 15 |
| Savings in % | 89 | 100 | 60 | 83 |
Summary
Automated culture using the BACT/ALERT 3D Microbial Detection System to screen for microbial contamination of platelet concentrates has been well documented over the past several decades. There have been numerous mitigation strategies that have been implemented to decrease contamination and improve detection including enhanced primary and secondary culture methods. Today there is a focus on platelet/patient safety and operational efficiency. Several models have been published on the economic impact of available testing strategies, and while one size doesn’t fit all the use of LVDS with 7-day shelf-life platelets has reported both improved safety and inventory management with a positive economic benefit.
While the test methodology has shown many changes over the past several decades, there have been only a few changes to the automated culture bottles (liquid emulsion sensor and plastic bottles) and the instrumentation has only recently evolved with the introduction of the BACT/ALERT VIRTUO. The BTA VIRTUO has a new instrument architecture to improve temperature stability; automation of loading and unloading culture bottles to improve workflow; and an enhanced detection algorithm that decreases time to detection of positive cultures.
Internal and external performance studies comparing the BTA VIRTUO and BTA 3D systems showed that the rate of detection and recovery was equivalent. The BTA VIRTUO did show improved performance over BTA 3D of between 2.1 to 2.8 hours faster in time to detection of contamination.
The specificity and sensitivity for the BTA VIRTUO was determined. A total of 485 negative controls were performed and no false positive (instrument positive, subculture negative) results were recorded. Similarly, of the 1,786 seeded bottles no false negative (instrument negative, subculture positive) results were recorded.
A time and motion study comparing the two systems showed that the BTA VIRTUO provided a 30% savings in footprint, 82% total savings in productivity and efficiency and 83% savings in time. The BTA VIRTUO system has a greater productivity advantage due to automated conveyor belt loading, scanning, and placement of the bottles within the system. This reduces human error and removes significant non-value-added tasks allowing the user to perform other functions within the laboratory.
With the new BTA VIRTUO system, the earlier detection of contaminants can lead to more timely response in recalling product or monitoring patients if the platelets have already been transfused. Additionally, with the improved laboratory efficiency, the system may show improved operational and economic benefits.
Acknowledgments
The authors thank Anne Hutchins and Virginie Le Coent for reviewing manuscript.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Sandra Ramirez-Arcos) for the series “Bacterial Contamination of Platelet Components” published in Annals of Blood. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aob-21-16). The series “Bacterial Contamination of Platelet Components” was commissioned by the editorial office without any funding or sponsorship. All authors are employees of bioMérieux. Ms. JY reports participation in a company share program. Mr. MU discloses he holds a patent for method and system for detection of microbial growth in a specimen container. The authors have no other conflicts of interests to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Blajchman MA, Beckers EAM, Dickmeiss E, et al. Bacterial detection of platelets: current problems and possible resolutions. Transfus Med Rev 2005;19:259-72. [Crossref] [PubMed]
- Levy JH, Neal MD, Herman JH. Bacterial contamination of platelets for transfusion: strategies for prevention. Crit Care 2018;22:271. [Crossref] [PubMed]
- McDonald CP. Bacterial risk reduction by improved donor arm disinfection, diversion and bacterial screening. Transfus Med 2006;16:381-96. [Crossref] [PubMed]
- Ramirez-Arcos S, Alport T, Goldman M. Intermittent bacteremia detected in an asymptomatic apheresis platelet donor with repeat positive culture for Escherichia coli: a case report. Transfusion 2015;55:2606-8. [Crossref] [PubMed]
- Hillyer CD, Josephson CD, Blajchman MA, et al. Bacterial contamination of blood components: risks, strategies, and regulation. Hematology Am Soc Hematol Educ Program 2003;575-89. [Crossref] [PubMed]
- Liumbruno GM, Catalano L, Piccinini V, et al. Reduction of the risk of bacterial contamination of blood components through diversion of the first part of the donation of blood and blood components. Blood Transfus 2009;7:86-93. [PubMed]
- Arpi M, Bremmelgaard A, Abel Y, et al. A novel screening method for the detection of microbial contamination of platelet concentrates: an experimental pilot study. Vox Sang 1993;65:335-6. [Crossref] [PubMed]
- Gong J, Högman CF, Lundholm M, et al. Novel automated microbial screening of platelet concentrates. APMIS 1994;102:72-8. [Crossref] [PubMed]
- Högman CF, Gong J. Studies of one invasive and two noninvasive methods for detection of bacterial contamination of platelet concentrates. Vox Sang 1994;67:351-5. [Crossref] [PubMed]
- Blajchman MA, Ali AM, Ricardson HL. Bacterial contamination of cellular blood components: risks, sources and control. Vox Sang 1994;67:25-33. [Crossref] [PubMed]
- Benjamin RJ, McDonald CP. The international experience of bacterial screen testing of platelet components with an automated microbial detection system: a need for consensus testing and reporting guidelines. Transfus Med Rev 2014;28:61-71. [Crossref] [PubMed]
- Prax M, Bekeredjian-Ding I, Krut O. Microbiological screening of platelet concentrates in Europe. Transfus Med Hemother 2019;46:76-86. [Crossref] [PubMed]
- Thorpe TC, Wilson ML, Turner JE, et al. BacT/Alert: an automated colorimetric microbial detection system. J Clin Microbiol 1990;28:1608-12. [Crossref] [PubMed]
- Wagner SJ, Robinette D. Evaluation of an automated microbiologic blood culture device for detection of bacteria in platelet components. Transfusion 1998;38:674-9. [Crossref] [PubMed]
- Liu HW, Yuen KY, Cheng TS, et al. Reduction of platelet transfusion-associated sepsis by short-term bacterial culture. Vox Sang 1999;77:1-5. [Crossref] [PubMed]
- Mertens G, Muylle L. False-positive and false-negative results of sterility testing of stored platelet concentrates. Transfusion 1999;39:539-40. [Crossref] [PubMed]
- Ollgaard M, Albjerg L, Georgsen J. Monitoring of bacterial growth in platelet concentrates - one year’s experience with the BactAlert system Vox Sang 1998;74:1126. (abstract).
- Laan E, Tros C. Improved safety and extended shelf-life of leuco-depleted platelet concentrates by automated bacterial screening Transfusion 1999;39:5S. (abstract).
- McDonald CP, Roy A, Lowe P, et al. Evaluation of the BacT/Alert automated blood culture system for detecting bacteria and measuring their growth kinetics in leucodepleted and non-leucodepleted platelet concentrates. Vox Sang 2001;81:154-60. [Crossref] [PubMed]
- Brecher ME, Means N, Jere CS, et al. Evaluation of an automated culture system for detecting bacterial contamination of platelets: an analysis with 15 contaminating organisms. Transfusion 2001;41:477-82. [Crossref] [PubMed]
- Brecher ME, Heath DG, Hay SN, et al. Evaluation of a new generation of culture bottle using an automated culture system for detecting nine common contaminating organisms found in platelet components. Transfusion 2002;42:774-9. [Crossref] [PubMed]
- Brecher ME, Hay SN, Rothenberg SJ. Evaluation of a new generation of plastic culture bottles with an automated microbial detection system for nine common contaminating organisms found in PLT components. Transfusion 2004;44:359-63. [Crossref] [PubMed]
- Fang CT, Chambers LA, Kennedy J, et al. Detection of bacterial contamination in apheresis platelet products: American Red Cross experience, 2004. Transfusion 2005;45:1845-52. [Crossref] [PubMed]
- Kleinman SH, Kamel HT, Harpool DR, et al. Two-year experience with aerobic culturing of apheresis and whole blood-derived platelets. Transfusion 2006;46:1787-94. [Crossref] [PubMed]
- Ramírez-Arcos S, Jenkins C, Dion J, et al. Canadian experience with detection of bacterial contamination in apheresis platelets. Transfusion 2007;47:421-9. [Crossref] [PubMed]
- Eder AF, Kennedy JM, Dy BA, et al. Bacterial screening of apheresis platelets and the residual risk of septic transfusion reactions: The American Red Cross experience (2004-2006). Transfusion 2007;47:1134-42. [Crossref] [PubMed]
- Benjamin RJ, Kline L, Dy BA, et al. Bacterial contamination of whole blood-derived platelets: the introduction of sample diversion and prestorage pooling with culture testing in the American Red Cross. Transfusion 2008;48:2348-55. [Crossref] [PubMed]
- Eder AF, Kennedy JM, Dy BA, et al. Limiting and detecting bacterial contamination of apheresis platelets: inlet-line diversion and increased culture volume improve component safety. Transfusion 2009;49:1554-63. [Crossref] [PubMed]
- Benjamin RJ, Dy B, Warren R, et al. Skin disinfection with a single-step 2% chlorhexidine swab is more effective than a two-step povidone-iodine method in preventing bacterial contamination of apheresis platelets. Transfusion 2011;51:531-8. [Crossref] [PubMed]
- Souza S, Bravo M, Poulin T, et al. Improving the performance of culture-based bacterial screening by increasing the sample volume from 4 mL to 8 mL in aerobic culture bottles. Transfusion 2012;52:1576-82. [Crossref] [PubMed]
- Bruhn R, Custer B, Vanderpool S, et al. Impact of increasing sample volume from 4 ml to 8 ml on bacterial detection rates in apheresis platelets: A meta-analysis. Vox Sang 2015;108:318-20. [Crossref] [PubMed]
- Ramírez-Arcos S, Kou Y, Mastronardi C, et al. Bacterial screening of outdated buffy coat platelet pools using a culture system and a rapid immunoassay. Transfusion 2011;51:2566-72. [Crossref] [PubMed]
- Jenkins C, Ramírez-Arcos S, Goldman M, et al. Bacterial contamination in platelets: incremental improvements drive down but do not eliminate risk. Transfusion 2011;51:2555-65. [Crossref] [PubMed]
- Ramirez-Arcos S, DiFranco C, McIntyre T, et al. Residual risk of bacterial contamination of platelets: six years of experience with sterility testing. Transfusion 2017;57:2174-81. [Crossref] [PubMed]
- de Korte D. 10 years experience with bacterial screening of platelet concentrates in the Netherlands. Transfus Med Hemother 2011;38:251-4. [Crossref] [PubMed]
- Borosak M, Wood E. Bacterial pre-release testing of platelets-the Australian Red Cross Blood Service clinical experience. Transfus Med Hemother 2011;38:239-41. [Crossref] [PubMed]
- Thyer J, Perkowska-Guse Z, Ismay SL, et al. Bacterial testing of platelets - has it prevented transfusion-transmitted bacterial infections in Australia? Vox Sang 2018;113:13-20. [Crossref] [PubMed]
- Murphy WG, Foley M, Doherty C, et al. Screening platelet concentrates for bacterial contamination: low numbers of bacteria and slow growth in contaminated units mandate an alternative approach to product safety. Vox Sang 2008;95:13-9. [Crossref] [PubMed]
- Bloch EM, Marshall CE, Boyd JS, et al. Implementation of secondary bacterial culture testing of platelets to mitigate residual risk of septic transfusion reactions. Transfusion 2018;58:1647-53. [Crossref] [PubMed]
- Tomasulo PA, Wagner SJ. Predicting improvement in detection of bacteria in apheresis platelets by maintaining constant component sampling proportion. Transfusion 2013;53:835-42. [Crossref] [PubMed]
- Kamel H, Townsend M, Bravo M, et al. Improved yield of minimal proportional sample volume platelet bacterial culture. Transfusion 2017;57:2413-9. [Crossref] [PubMed]
- McDonald C, Allen J, Brailsford S, et al. Bacterial screening of platelet components by National Health Service Blood and Transplant, an effective risk reduction measure. Transfusion 2017;57:1122-31. [Crossref] [PubMed]
- Delage G, Bernier F, Beaudoin J, et al. Improved bacterial culture of platelet product: preliminary results after implementation of a two-bottle system with 48-hour sampling Transfusion 2016;56:28A. (abstract).
- Bernier F, Delage G. Improving the safety of platelets by increasing the sample volume and time for sampling and issuing - bacteriological results Vox Sang 2018;113:2018. (abstract).
- Blood Products Advisory Committee November 30- December 1, 2017 Meeting Announcement - 11/30/2017 - 12/01/2017 | FDA [Internet]. [cited 2020 Dec 21]. Available online: https://www.fda.gov/advisory-committees/advisory-committee-calendar/blood-products-advisory-committee-november-30-december-1-2017-meeting-announcement-11302017-12012017
- Blood Products Advisory Committee July 18-19, 2018 Meeting Announcement - 07/17/2018 - 07/18/2018 | FDA [Internet]. [cited 2020 Dec 21]. Available online: https://www.fda.gov/advisory-committees/advisory-committee-calendar/blood-products-advisory-committee-july-18-19-2018-meeting-announcement-07172018-07182018
- Walker BS, White SK, Schmidt RL, et al. Residual bacterial detection rates after primary culture as determined by secondary culture and rapid testing in platelet components: a systematic review and meta-analysis. Transfusion 2020;60:2029-37. [Crossref] [PubMed]
- White SK, Schmidt RL, Walker BS, et al. Bacterial contamination rate of platelet components by primary culture: a systematic review and meta-analysis. Transfusion 2020;60:986-96. [Crossref] [PubMed]
- Jones SA, Jones JM, Leung V, et al. Sepsis attributed to bacterial contamination of platelets associated with a potential common source - multiple states, 2018. MMWR Morb Mortal Wkly Rep 2019;68:519-23. [Crossref] [PubMed]
- Bacterial Risk Control Strategies for Blood Collection Establishments and Transfusion Services to Enhance the Safety and Availability of Platelets for Transfusion | FDA [Internet]. [cited 2020 Dec 21]. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/bacterial-risk-control-strategies-blood-collection-establishments-and-transfusion-services-enhance
- BK200472 - BACT/ALERT BPA Culture Bottle; BACT/ALERT BPN Culture Bottle | FDA [Internet]. [cited 2020 Dec 21]. Available online: https://www.fda.gov/vaccines-blood-biologics/substantially-equivalent-510k-device-information/bk200472-bactalert-bpa-culture-bottle-bactalert-bpn-culture-bottle
- Blake JT. Determining the inventory impact of extended-shelf-life platelets with a network simulation model. Transfusion 2017;57:3001-8. [Crossref] [PubMed]
- Lu W, Delaney M, Flegel WA, et al. How do you… decide which platelet bacterial risk mitigation strategy to select for your hospital-based transfusion service? Transfusion 2020;60:675-81. [Crossref] [PubMed]
- Janssen MP, Van Der Poel CL, Buskens E, et al. Costs and benefits of bacterial culturing and pathogen reduction in the Netherlands. Transfusion 2006;46:956-65. [Crossref] [PubMed]
- McCullough J, Goldfinger D, Gorlin J, et al. Cost implications of implementation of pathogen-inactivated platelets. Transfusion 2015;55:2312-20. [Crossref] [PubMed]
- Li JW, Brecher ME, Jacobson JL, et al. Addressing the risk of bacterial contamination in platelets: a hospital economic perspective. Transfusion 2017;57:2321-8. [Crossref] [PubMed]
- Prioli KM, Karp JK, Lyons NM, et al. Economic implications of pathogen reduced and bacterially tested platelet components: a US hospital budget impact model. Appl Health Econ Health Policy 2018;16:889-99. [Crossref] [PubMed]
- Kacker S, Katz LM, Ness PM, et al. Financial analysis of large-volume delayed sampling to reduce bacterial contamination of platelets. Transfusion 2020;60:997-1002. [Crossref] [PubMed]
- Lu W, Fung M. Platelets treated with pathogen reduction technology: Current status and future direction. F1000Research 2020;9:1-8. [Crossref] [PubMed]
- Lu W, Delaney M, Dunbar NM, et al. A national survey of hospital-based transfusion services on their approaches to platelet bacterial risk mitigation in response to the FDA final guidance for industry. Transfusion 2020;60:1681-7. [Crossref] [PubMed]
- Savinkina AA, Haass KA, Sapiano MRP, et al. Transfusion-associated adverse events and implementation of blood safety measures - findings from the 2017 National Blood Collection and Utilization Survey. Transfusion 2020;60:S10-6. [Crossref] [PubMed]
- Earnshaw SR, Beyhaghi H, McDade C, et al. Economic evaluation of large-volume delayed sampling and pathogen reduction technology strategies for processing platelets Transfusion 2020;60:39A-40A. (abstract).
- Ramirez‐Arcos S, Evans S, McIntyre T, et al. Extension of platelet shelf life with an improved bacterial testing algorithm. Transfusion 2020;60:2918-28. [Crossref] [PubMed]
- Paris A, Deol P, Adamik M, et al. Rapid detection of bacterial contaminants in platelet components: comparison of time to detection between the Bact/Alert® VirtuoTM and the Bact/Alert® 3D Vox Sang 2016;111:196. (abstract).
- Allen J. BacT/ALERT Virtuo: a new generation for bacterial screening. In: British Blood Transfusion Society 2018. Available online: https://www.bbts.org.uk/bbts2018/programme1/2018presentations/
- Jacobs M, Good CE, Allen J, et al. Multicenter comparison of the BacT/ALERT VIRTUO and BacT/ALERT 3D instruments for the rapid detection of bacterial contaminants in leukocyte reduced apheresis platelets Transfusion 2017;57:195A. (abstract).
- Adamik M, Viray J, Helms D, et al. Rapid detection of microbial contamination in leukocyte reduced apheresis platelets during five day expiry: repeatability testing on the BacT/ALERT® VIRTUO and the BacT/ALERT® 3D Transfusion 2017;57:200A-201A. (abstract).
- Daane L, Ahmed M, Yang J, et al. Automated microbial detection system time and motion study: BACT/ALERT®3D versus VIRTUO® Transfusion 2019;59:49A-50A. (abstract).
Cite this article as: Daane L, Adamik M, Ullery M, Ahmed M, Yang J. Rapid detection of bacterial contaminants in platelet concentrates using the next generation BACT/ALERT® VIRTUO® Microbial Detection System: improved safety and operational efficiency. Ann Blood 2021;6:40.