Molecular typing of blood group genes in diagnostics
Introduction
Blood group antigens are antigenic determinants on the surface of red blood cells (RBCs). That is, they can elicit an immune response after a transfusion or pregnancy event. It is the antibody that causes clinical problems in transfusion incompatibility, maternal-fetal incompatibility, and autoimmune hemolytic anemia (1).
Alloimmunization is the source of a variety of problems during long-term medical and transfusion management, with the main problems being the correct definition of many clinically significant antigens and the identification of appropriate antigen-negative RBCs for transfusion (2-4). At the inception of immunohematology and for decades after, hemagglutination test results were used to determine the phenotype and to predict the genotype of an individual. Despite its relatively low cost, ease of performance, sensitivity, and specificity, hemagglutination-based determination of RBC phenotype has important limitations. Accurate antigen typing of transfused patients is often a difficult task due to the presence of donor RBCs in the patients’ circulation. It is also complicated to type cells when a patient’s RBCs have a positive direct antiglobulin test and no direct agglutinating antibody is available and there are no commercial antibody reagents to type many clinically significant antigens. In addition, the lack of automation in hemagglutination may be a limiting factor in the routine typing of blood group antigens.
In part, to mitigate these limitations, molecular typing of blood group genes has been added in diagnostics and is increasingly being used to determine the genotype and to predict the phenotype of an individual (5,6).
Table 1 shows the advantages of molecular typing in diagnostics compared to hemagglutination.

Full table
Applications of molecular typing of blood group genes in diagnostics
The use of the genetic information provided by the Human Genome Project added a new phase in immunohematology. Molecular bases of the 343 blood group antigens clustered in 43 blood group systems now recognized by International Society of Blood Transfusion (ISBT) were elucidated (7,8). With the knowledge gathered with the gene cloning and sequencing of blood group genes, it was possible to identify the molecular characteristics of the blood group antigens and to know that most of them are derived of single nucleotide variations (SNVs), know the RH locus organization and developing a multitude of methods for blood group phenotyping using DNA-based technology (9-16). And then several advantages of molecular methods over serological methods in diagnostics appeared. It was shown that we do not need RBCs for RBC genotyping as genomic DNA can be obtained by different sources of cells and that transfused cells do not interfere and therefore we could test patients who have being recently transfused (17).It was also demonstrated that molecular tests could be used to type patients with warm autoantibodies, type low and high frequency antigens, assist in the identification of variant antigens and in the differentiation between weak D and partial D (18).
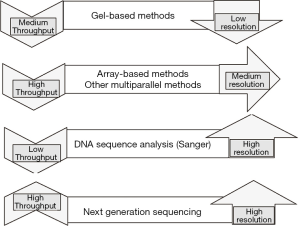
Table 2 summarizes the main applications of molecular typing in diagnostics and Figure 1 shows the molecular based methods currently applied in diagnostics in terms of resolution and throughput.
Molecular discoveries brought a lot of other benefits to Transfusion Medicine including the discoveries of new blood group systems and the possibility of performing personalized therapy. First, new technology since monoclonal antibodies in 1980s with development of single nucleotide polymorphism (SNP) arrays and next generation sequencing (NGS) provided us with new blood group genes and proteins and the assignment of orphan antigens to their blood group home.
Molecular typing in patient diagnostics
Molecular typing of blood group genes in the transfusion setting is recommended for transfusion-dependent patients, as part of antibody identification process, since the identification of the patient’s predicted phenotype allows the laboratory to determine to which antigens the patient can and cannot respond to make alloantibodies. In addition, molecular typing provides improved accuracy and more information on the antigenic profile of the patients, especially those where no serological reagents are available as for example the Dombrock (DO) antigens and rare blood types.
Several studies have shown the relevance of blood group genotyping for the management of chronically transfused patients with diseases such as sickle cell anemia (SCA), thalassemia and myelodysplastic syndrome (MDS), in which the hemagglutination results may not reflect the patient’s true phenotype, by assisting in the identification of suspected alloantibodies and the selection of antigen-negative RBCs for transfusion (21-24). Taking the genotype into account enables a better selection of compatible units for patients and clinical benefits for them. Several authors showed that blood group genotyping of transfusion-dependent patients is useful in preventing and identifying alloimmunization and in decreasing the risk of hemolytic transfusion reactions (25). Also, the use of better-matched blood units can reduce transfusion requirements, decreasing the risk of other adverse reactions like transfusion-related acute lung injury and potential exposure to infectious diseases.
Sickle cell patients are among those who benefit most from molecular typing as it allows for more extended antigen matching, identifies patients who lack high-prevalence antigens and helps to differentiate between auto- and alloantibodies, especially in patients who make Rh antibodies despite D, C, and E antigen matching by serology. In a previous study, da Costa et al. (26) demonstrated that molecular matching of red cells is superior to serological matching in patients with sickle cell disease (SCD) when patients who were receiving serologic matching units, received precisely genotypically-matched units and benefited of the transfusions as shown by better in vivo RBCs survival, as assessed by raises in hemoglobin levels and diminished frequency of transfusions. The use of the DNA approach was also essential to identify autoantibodies underlying clinically significant alloantibodies.
The great diversity of RH genes in African individuals also increases the risk of patients with SCD to develop clinically significant Rh antibodies (27-29). Chou et al. (30) reported RH genotypes in 857 patients with SCD showing that 29% of RHD and 53% of RHCE were altered, but not all antibodies developed by those patients were associated with inheritance of altered RH alleles. These results suggest that Rh antibodies are not only a result of inheriting altered RH alleles, but may also be a result of altered Rh epitopes on donor red cells. Recently, Macedo et al. (31) provided evidence that patients exposed to RBC units from donors with Rh variants may develop clinically significant Rh antibodies. Therefore, molecular typing can assist in the identification of RHD and RHCE alleles encoding altered Rh epitopes in patients and blood donors, improving RH genotype-matched RBC units and providing the means for reducing Rh alloimmunization and delayed hemolytic transfusion reactions (DHTRs).
In addition, the discrimination between weak D and partial D in patients with SCD can be of clinical importance, because carriers of partial D antigen may develop anti-D when transfused with D-positive RBC units. For example, a systematic RHD genotyping in 48 Brazilian patients with SCD serologically-typed as weak D showed that 40 of them had RHD genes encoding partial D and four patients had already developed anti-D, demonstrating the importance to differentiate weak D and partial D in chronically transfused patients to establish a transfusion policy recommendation (32).
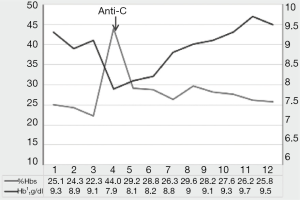
Besides, the knowledge of the specific RH variant inherited or exposed is important to predict the clinical significance of Rh antibodies produced by patients with SCD. Coleman et al. (33) found that among alloimmunized patients with SCD receiving D, CEK antigen-matched RBCs, 52.3% had Rh specificities, of which 26.5% associated with specific variant alleles had DHTR. In a recent study, Miranda et al. (34) evaluated eleven patients with SCD with Rh alloantibodies and Rh variants receiving serologic Rh-matched RBC units, and verified that seven patients had laboratory and clinical evidence of hemolysis due to anti-C produced by four patients with the RHCE*(C)ceS/RHCE*(C)ceS genotype and to anti-e produced by a patient with a homozygous RHCE*ceAR allele. Figure 2 shows transfusion follow-up of one of the patients RHCE*(C)ceS homozygous who developed anti-C after transfusion of a C+ RBC unit.
In addition to its contribution to the general accuracy of identification of RBC antigens and variants, genotyping of transfusion-dependent patients with SCD is important to determine which phenotypically Fy(b−) patients can safely receive Fy(b+) RBCs without the risk of alloimmunization (35). The use of FY genotyping for identification of patients with SCD carrying the FY*B-67C allele has been shown to considerably increase the availability of blood for them.
Patients with warm autoantibodies or with drug interference (drug antibodies or monoclonal antibody drug therapies such as anti-CD38 and anti-CD47) have also benefited from extended RBC genotype with the possibility of receiving transfusions of RBC units matched to clinically significant antigens (36-38). This approach reduces the risk of hemolytic transfusion reactions, prevents further alloimmunization and improves patient care by reducing working time and the number of tests performed to remove the interference of autoantibodies or drugs underlying RBC alloantibodies.
Molecular typing in donor testing
Molecular typing can be used to antigen-type blood donors for transfusion, as multiple SNVs can be included in a single assay, allowing efficient screening for multiple antigens. Currently, high-throughput genotyping based on DNA arrays is a very feasible method to obtain a fully typed donor database to be used for a better matching between recipient and donor to prevent alloimmunization and hemolytic transfusion reactions. In addition, it is expected to greatly expand the pool of blood donors who are negative for multiple antigens or negative for a high-prevalence antigen (39,40).
Molecular typing also provides the means to identify antigen-negative donors when typing antisera are not available or are weakly reactive, e.g., anti-Doa, -Dob, -Hy, -Joa, -V, -VS, and to provide a better characterization of reagent cell panels that are used for patient antibody identification (41).
Another significant advantage of molecular typing in blood donors is the identification of variant alleles leading to weak antigens expression as they can immunize an antigen-negative transfusion recipient if not recognized. An example is a donor with the Fyx phenotype, who may be mistyped as Fy(b−) by serology. Molecular typing can correctly characterize this donor as Fy(b+w) by identification of the 265C>T SNV responsible for the weak antigen expression. In a recent study reporting internal discrepancies detected when testing a subsequent donation by DNA testing the authors showed that 52% of the discrepancies were associated with the FY system, and most involved the Fyx phenotype (42).
The molecular typing of a variant gene can also assist in resolving a serologic investigation in blood donors. A proportion of donors with ABO subgroups who have been typed as group O in the past are now being recognized as group A or group B with the use of monoclonal antibodies. Since the bases of many of the weak subgroups of A and B are associated with altered transferase genes, DNA-based assays can be used to correctly define the ABO group of those donors (43). Similarly, a proportion of blood donors are presenting discrepancies in D typing due to the variability to detect weak and partial antigens (44,45). Molecular testing has been used to confirm the RhD type of donors in order to identify donors with the potential to alloimmunize RhD-negative recipients. A recent study performed in the United States in 1,174 serologic RhD-negative blood donors showed that 0.94% had variant RHD alleles that might cause alloimmunization in RhD-negative recipients (46).
Molecular typing in prenatal testing
The discovery of cell-free fetal DNA (cffDNA) in maternal plasma has opened new and exciting possibilities for the non-invasive prenatal determination of fetal blood group status following fetal-maternal bleeding (47). Non-invasive genotyping of fetal RHD status by analyzing cffDNA in maternal plasma has already been incorporated into routine practice of many countries for the management of D-negative pregnant women previously sensitized or at risk of immunization (48,49). The assessment of the risk of hemolytic disease of the fetus and newborn (HDFN) by accurate prediction of fetal RHD status allows to administrate Rh immunoglobulin (RhIg) to D-negative women with D-positive fetuses and avoids unnecessary administration in cases of D-negative fetuses (50).
Appropriate classification of RhD phenotypes is also critical to determining the management of obstetric patients (51,52). However, the serologic distinction between partial D and weak D is a difficult task due to the variations in serologic testing (53). RHD genotyping has been used to provide information about risk of alloimmunization and candidacy for RhIg. It is generally accepted that women who have a serologic weak D phenotype due to RHD*weak D types 1, 2, and 3 alleles, the most common RhD variants in Caucasians, are not at risk of alloimmunization and can be treated as RhD-positive avoiding the unnecessary use of RhIg (54,55). According to Sandler et al. (54), around 13,360 pregnant women whose RhD type was uncertain, received RhIg when it was not necessary, which means an estimated 26,700 doses administered to patients who did not need it. However, particular RHD variant alleles are known to be more prevalent in different ethnic groups. African descent individuals are more likely to express a partial D phenotype (32,56). In a previous study, Bub et al. (57) showed a great variability in RHD variant alleles in pregnant women from a population of high admixture. According to the results obtained, 78% of these obstetric patients were at risk for anti-D and candidates for RhIg.
Based on the prevalence of RHD variant alleles in different ethnic groups, there is a consensus in the literature that pregnant women with weak D phenotype should be provided with RHD genotyping for determination of weak or partial D status in order to identify women who are candidates for RhIg. And this analysis has demonstrated be clinically beneficial without increasing overall costs (58).
Cost-effectiveness of molecular typing in diagnostics
The cost and reimbursement of molecular tests has been a barrier to their routine implementation. However, the cost of molecular detection can be significantly reduced by analyzing several polymorphisms at the same time and processing a large number of samples per reaction using a centralized testing model. The introduction of short-read NGS technologies has also led to a reduction in sequencing costs. It is important to note that molecular testing would need to be performed only once per lifetime and can make part of the patient transfusion record. There is currently a consensus that molecular typing is cost-efficient to select more compatible blood for transfusion facilitating transfusion, preventing, and reducing RBC alloimmunization and DHTR (59). In addition, it can reduce the cost of Rh prophylaxis, discomfort from Ig application and exposure to infectious agents. In repeat donors the use of molecular typing can reduce the cost of repetitions and in the acquisition of rare sera with a probability of achieving more accurate compatibility. On the other hand, it has been demonstrated that even though the hemagglutination tests are considered faster and cheaper than molecular tests, for patients with many specific diagnoses and serologic and transfusion history characteristics, they can be limited, expensive and time-consuming. Cost-benefit analysis is often challenging, but the costs of an alloimmunized patient and a transfusion reaction must be taking into account when analyzing whether the molecular typing of blood group genes in diagnostic is cost-saving or cost-effective. Target group of patients, such as those with SCD or with autoimmune hemolytic anemia and those with unexplained complex serological results, will particularly benefit from molecular assays with more efficient, faster, and less costly analysis, improving safety and efficacy of blood products with low additional costs.
Summary
Molecular typing of blood group genes brings a new era in, diagnostics offering many advantages over serologic testing, with the primary benefit of predicting the blood group phenotype in situations that cannot be performed serologically. It has been successfully implemented in clinical laboratories and is proving to be a powerful tool, preventing alloantibodies formation, with potential advantages for identifying rare blood types and finding better antigen matches for chronically transfused patients.
Advances are ongoing in the automation of SNVs and DNA sequence analysis and the success of sequencing the human genome has shown the potential for genotyping large numbers of samples. With the advance of NGS, it will be possible obtain accurate blood group phenotype predictions in both blood donors and patients and bring personalized medicine to the field of Transfusion Medicine (60-63) by allowing a more precise red cell matching to the patients. This would change current practice, increasing the inventories of phenotyped RBC units, enabling blood transfusions with a superior match for women in childbearing age and for patients requiring chronic transfusions, thus preventing immunization, the risk of HDFN and hemolytic transfusion reactions.
Hemagglutination has identified many phenotypic variants encoded by RHD, RHCE, or hybrids, and molecular analysis has revealed remarkable variation within the variants. Numerous partial D and weak D phenotypes have been defined at the molecular level (8) and this information, together with clinical and serologic data, has been used to guide transfusion and testing policy for patients and donors.
The wide availability of serologically-defined variants has contributed to the knowledge about the allelic diversity of the human blood groups. Although this diversity of blood group alleles is not always associated with adverse transfusion reactions, with the gathering of more information it became clear that many molecular events result in discrepancies between phenotype and genotype. In a recent study (64) evaluating 325 discrepancies found between phenotypes and genotypes in daily routine practice, the authors verified that 97.67% of the discrepancies occurred due to false phenotype results in hemagglutination, demonstrating that despite the limitations of molecular methods currently employed (Table 3), genotyping is more efficient to define the blood types, especially in transfusion dependent patients.

Full table
The knowledge gathered with molecular discoveries has also been used to explore expression of new alleles, express antigens in heterologous systems by using mRNA in transfection studies to potentially detect and identify blood group antibodies in a single automated assay and to produce recombinant forms of an antigen to facilitate the detection of alloantibodies to high-prevalence antigens.
The advance of NGS tests with the inclusion of genetic variants leading to null phenotypes and the implementation in the clinical setting with low additional costs, will overcome the limitations of the current molecular methods and may allow the full replacement of blood group phenotyping in a near future.
Acknowledgments
The author would like to thank Tamires Delfino dos Santos, Maria Rita Miranda and Simone Gilli from Hemocentro Unicamp for their laboratory and clinical support.
Funding: This study was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) grant No. 2020/02191-1.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Yann Fichou) for the series “Molecular Genetics and Genomics of Blood Group Systems” published in Annals of Blood. The article has undergone external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aob-20-73). The series “Molecular Genetics and Genomics of Blood Group Systems” was commissioned by the editorial office without any funding or sponsorship. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Poole J, Daniels G. Blood group antibodies and their significance in transfusion medicine. Transfus Med Rev 2007;21:58-71. [Crossref] [PubMed]
- Zimring JC, Welniack L, Semple JW, et al. Current problems and future directions of transfusion-induced alloimmunization: summary of an NHLBI working group. Transfusion 2011;51:435-41. [Crossref] [PubMed]
- Schonewille H, van de Watering LM, Brand A. Additional red blood cell alloantibodies after blood transfusions in a nonhematologic patient cohort: is it time to take precautionary measures? Transfusion 2006;46:630-5. [Crossref] [PubMed]
- Chou ST, Liem RI, Thompson AA. Challenges of alloimmunization in patients with haemoglobinopathies. Br J Haematol 2012;159:394-404. [Crossref] [PubMed]
- Denomme GA, Flegel WA. Applying molecular immunohematology discoveries to standards of practice in blood banks: now is the time. Transfusion 2008;48:2461-75. [Crossref] [PubMed]
- Hillyer CD, Shaz BH, Winkler AM, et al. Integrating molecular technologies for red blood cell typing and compatibility testing into blood centers and transfusion services. Transfus Med Rev 2008;22:117-32. [Crossref] [PubMed]
- Storry JR, Clausen FB, Castilho L, et al. International Society of Blood Transfusion Working Party on Red Cell Immunogenetics and Blood Group Terminology: Report of the Dubai, Copenhagen and Toronto meetings. Vox Sang 2019;114:95-102. [Crossref] [PubMed]
ISBT-Working Party on Red Cell Immunogenetics and Blood Group Terminology - Denomme GA. Molecular basis of blood group expression. Transfus Apher Sci 2011;44:53-63. [Crossref] [PubMed]
- Reid ME, Denomme GA. DNA-based methods in the immunohematology reference laboratory. Transfus Apher Sci 2011;44:65-72. [Crossref] [PubMed]
- Boccoz SA, Le Goff G, Blum LJ, et al. Microarrays in blood group genotyping. Molecular typing of blood cell antigens. Methods Mol Biol 2015;1310:105-13. [Crossref] [PubMed]
- Meyer S, Vollmert C, Trost N, et al. High-throughput Kell, Kidd, and Duffy matrix-assisted laser desorption/ionization, time-of-flight mass spectrometry-based blood group genotyping of 4000 donors shows close to full concordance with serotyping and detects new alleles. Transfusion 2014;54:3198-207. [Crossref] [PubMed]
- Latini FR, Gazito D, Arnoni CP, et al. A new strategy to identify rare blood donors: single polymerase chain reaction multiplex SNaPshot reaction for detection of 16 blood group alleles. Blood Transfus 2014;12:s256-63. [PubMed]
- Wagner FF, Bittner R, Döscher A, et al. Extended Donor Typing by Pooled Capillary Electrophoresis: Impact in a Routine Setting. Transfus Med Hemother 2018;45:225-37. [Crossref] [PubMed]
- Fichou Y, Audrézet MP, Guéguen P, et al. Next-generation sequencing is a credible strategy for blood group genotyping. Br J Haematol 2014;167:554-62. [Crossref] [PubMed]
- Boccoz SA, Fouret J, Roche M, et al. Massively parallel and multiplex blood group genotyping using next-generation-sequencing. Clin Biochem 2018;60:71-6. [Crossref] [PubMed]
- Reid ME, Rios M, Powell D, et al. DNA from blood samples can be used to genotype patients who have recently received a transfusion. Transfusion 2000;40:48-53. [Crossref] [PubMed]
- Westhoff CM. Blood group genotyping. Blood 2019;133:1814-20. [Crossref] [PubMed]
- Kutner JM, Mota M, Conti F, et al. Blood genotyping for improved outcomes in chronic transfusion patients: current and future perspectives. Int J Clin Transfus Med 2014;2:65-72. [Crossref]
- Reid ME, Lomas-Francis C. Molecular approaches to blood group identification. Curr Opin Hematol 2002;9:152-9. [Crossref] [PubMed]
- Castilho L, Rios M, Bianco C, et al. DNA-based typing of blood groups for the management of multiply-transfused sickle cell disease patients. Transfusion 2002;42:232-8. [Crossref] [PubMed]
- Castilho L, Rios M, Pellegrino J Jr, et al. Blood group genotyping facilitates transfusion of beta-thalassemia patients. J Clin Lab Anal 2002;16:216-20. [Crossref] [PubMed]
- Ribeiro KR, Guarnieri MH, da Costa DC, et al. DNA array analysis for red blood cell antigens facilitates the transfusion support with antigen-matched blood in patients with sickle cell disease. Vox Sang 2009;97:147-52. [Crossref] [PubMed]
- Guelsin GAS, Fujita CR, Rodrigues C, et al. Molecular matching for Rh and K reduces red blood cell alloimmunization in patients with myelodisplastic syndrome. Blood Transfus 2015;13:53-8. [PubMed]
- Fasano RM, Chou S. Red Blood Cell Antigen Genotyping for Sickle Cell Disease, Thalassemia, and Other Transfusion Complications. Transfus Med Rev 2016;30:197-201. [Crossref] [PubMed]
- da Costa DC, Pellegrino J, Guelsin GAS, et al. Molecular matching of red blood cells is superior to serological matching in sickle cell disease patients. Rev Bras Hematol Hemoter 2013;35:35-8. [Crossref] [PubMed]
- Chou ST, Jackson T, Vege S, et al. High prevalence of red blood cell alloimmunization in sickle cell disease despite transfusion from Rh-matched minority donors. Blood 2013;122:1062-71. [Crossref] [PubMed]
- Dinardo CL, Kelly S, Dezan MR, et al. Diversity of RH and transfusion support in Brazilian sickle cell disease patients with unexplained Rh antibodies. Transfusion 2019;59:3228-35. [Crossref] [PubMed]
- Chou ST, Flanagan JM, Vege S, et al. Whole-exome sequencing for RH genotyping and alloimmunization risk in children with sickle cell anemia. Blood Adv 2017;1:1414-22. [Crossref] [PubMed]
- Chou ST, Evans P, Vege S, et al. RH genotype matching for transfusion support in sickle cell disease. Blood 2018;132:1198-207. [Crossref] [PubMed]
- Macedo MD, Miranda MR, Santos TD, et al. Rh antibodies as a result of altered Rh epitopes on transfused red cells: a case series of seven Brazilian patients. Blood Transfus 2020; [Crossref] [PubMed]
- Leal I, Santos TD, Miranda MR, et al. Systematic RHD Genotyping in Brazilians Reveals a High Frequency of Partial D in Chronically Transfused Patients Serologically Typed As Weak D. Hematol Transf Cell Ther 2020;42:348. [Crossref]
- Coleman S, Westhoff CM, Friedman DF, et al. Alloimmunization in patients with sickle cell disease and underrecognition of accompanying delayed hemolytic transfusion reactions. Transfusion 2019;59:2282-91. [Crossref] [PubMed]
- Miranda MR, Leal I, Santos TD, et al. Clinical Outcomes of Sickle Cell Disease Patients with Unexpected Rh Antibodies Receiving Serologic Rh-Matched Red Blood Cell Units. Hematol Transf Cell Ther 2020;42:339. [Crossref]
- Castilho L. The value of DNA analysis for antigens in the Duffy blood group system. Transfusion 2007;47:28S-31S. [Crossref] [PubMed]
- El Kenz H, Efira A, Quoc Le P, et al. Transfusion support of autoimmune hemolytic anemia: how could the blood group genotyping help? Transl Res 2014;163:36-42. [Crossref] [PubMed]
- Bub CB, Reis IN, Aravechia MG, et al. Transfusion management for patients taking an anti-CD38 monoclonal antibody. Rev Bras Hematol Hemoter 2018;40:25-9. [PubMed]
- Velliquette RW, Aeschlimann J, Kirkegaard J, et al. Monoclonal anti-CD47 interference in red cell and platelet testing. Transfusion 2019;59:730-7. [Crossref] [PubMed]
- Flegel WA, Gottschall JL, Denomme GA. Implementaing mass-scale red cell genotyping at a blood center. Transfusion 2015;55:2610-5. [Crossref] [PubMed]
- Shafi H, Abumuhor I, Klapper E. How we incorporate molecular typing of donors and patients into our hospital transfusion service. Transfusion 2014;54:1212-9. [Crossref] [PubMed]
- Scharberg E, Rink G, Portegys J, et al. The Impact of Using Genotyped Reagent Red Blood Cells in Antibody Identification. Transfus Med Hemother 2018;45:218-24. [Crossref] [PubMed]
- DePalma H, Godbey EA, Opalka A, et al. Reliability of labeling red cell units with minor antigen historical results and process considerations. Transfusion 2020;60:822-30. [Crossref] [PubMed]
- Olsson ML, Irshaid NM, Hosseini-Maaf B, et al. Genomic analysis of clinical samples with serologic ABO blood grouping discrepancies: identification of 15 novel A and B subgroup alleles. Blood 2001;98:1585-93. [Crossref] [PubMed]
- Wagner FF, Gassner C, Müller TH, et al. Molecular basis of weak D phenotypes. Blood 1999;93:385-93. [Crossref] [PubMed]
- Flegel WA, von Zabern I, Wagner FF. Six years’experience performing RGD genotyping to confirm D- red blood vell units in Germany for preventing anti-D immunizations. Transfusion 2009;49:465-71. [Crossref] [PubMed]
- Perez-Alvarez I, Hayes C, Hailemariam T, et al. RHD genotyping of serologic RhD-negative blood donors in a hospital-based blood donor center. Transfusion 2019;59:2422-8. [Crossref] [PubMed]
- Lo YMD. Fetal RhD genotyping from maternal plasma. Ann Med 1999;31:308-12. [Crossref] [PubMed]
- Clausen FB. Integration of noninvasive prenatal prediction of fetal blood group into clinical prenatal care. Prenat Diagn 2014;34:409-15. [Crossref] [PubMed]
- Clausen FB, Steffensen R, Christiansen M, et al. Routine noninvasive prenatal screening for fetal RHD in plasma of RhD-negative pregnant women-2 years of screening experience from Denmark. Prenat Diagn 2014;34:1000-5. [Crossref] [PubMed]
- Clausen FB, Rieneck K, Krog GR, et al. Noninvasive Antenatal Screening for Fetal RHD in RhD Negative Women to Guide Targeted Anti-D Prophylaxis. Methods Mol Biol 2019;1885:347-59. [Crossref] [PubMed]
- Denomme GA, Wagner FF, Fernandes BJ, et al. Partial D, weak D types, and novel RHD alleles among 33,864 multiethnic patients: implications for anti-D alloimmunization and prevention. Transfusion 2005;45:1554-60. [Crossref] [PubMed]
- Flegel WA, Denomme GA, Yazer MH. On the complexity of D antigen typing: a handy decision tree in te age of molecular blood group diagnostics J Obstet Gynaecol Can 2007;29:746-52. (Review). [Crossref] [PubMed]
- Denomme GA, Dake LR, Vilensky D, et al. Rh discrepancies caused by variable reactivity of partial and weak D types with different serologic techniques. Transfusion 2008;48:473-8. [Crossref] [PubMed]
- Sandler SG, Flegel WA, Westhoff CM, et al. It’s time to phase in RHD genotyping for patients with a serologic weak D phenotype. College of American Pathologists Transfusion Medicine Resource Committee Work Group. Transfusion 2015;55:680-9. [Crossref] [PubMed]
- Sandler SG, Chen LN, Flegel WA. Serological weak D phenotypes: a review and guidance for interpreting the RhD blood type using the RHD genotype. Br J Haematol 2017;179:10-9. [Crossref] [PubMed]
- Keller MA, Crowley J, Horn T, et al. Characterization of RH locus in African American Blood Donors. Transfusion 2013;53:28A.
- Bub CB, Aravechia MG, Costa TH, et al. RHD alleles among pregnant women with serologic discrepant weak D phenotypes from a multiethnic population and risk of alloimmunization. J Clin Lab Anal 2018;32:e22221 [Crossref] [PubMed]
- Kacker S, Vassallo R, Keller M, et al. Financial implications of RHD genotyping of pregnant women with a serologic weak D phenotype. Transfusion 2015;55:2095-103. [Crossref] [PubMed]
- Carter JH, Flegel WA. Red cell transfusions in genomic era. Semin Hematol 2019;56:236-40. [Crossref] [PubMed]
- Fichou Y, Ferec C. NGS and blood group systems: State of the art and perspectives. Transfus Clin Biol 2017;24:240-4. [Crossref] [PubMed]
- Lane WJ, Westhoff CM, Uy JM, et al. Comprehensive blood cell and platelet antigen prediction from whole genome sequencing: proof of principle. Transfusion 2016;56:743-54. [Crossref] [PubMed]
- Fichou Y, Mariez M, Le Marechal C, et al. The experience of extended blood group genotyping by next generation sequencing (NGS): investigation of patients with sickle cell disease. Vox Sang 2016;111:418-24. [Crossref] [PubMed]
- Guo Y, Busch MP, Seielstad M, et al. Development and evaluation of a transfusion medicine genome wide genotyping array. Transfusion 2019;59:101-11. [Crossref] [PubMed]
- Menegati SFP, Santos TD, Macedo MD, et al. Discrepancies between red cell phenotyping and genotyping in daily immunohematology laboratory practice. Transfus Apher Sci 2020;59:102585 [Crossref] [PubMed]
Cite this article as: Castilho L. Molecular typing of blood group genes in diagnostics. Ann Blood 2021;6:20.