
The PGDprime immunoassay for detection of bacterial contamination in platelet concentrates: sensitivity and specificity
Introduction
The PGDprime Test is a rapid, qualitative immunoassay for the detection of aerobic and anaerobic Gram-positive (GP) and Gram-negative (GN) bacteria (1). It is FDA cleared to detect bacteria in:
- Leukocyte-reduced apheresis platelets (LRAP) suspended in plasma, LRAP suspended in platelet additive solution (PAS-C) and plasma, and pre-storage pools (PSP) of up to six leukoreduced (LR) whole blood-derived platelets (WBDP) suspended in plasma, within 24 hours before platelet transfusion as a “safety measure” after testing with a growth-based quality control test cleared by the FDA for platelet components;
- Post-storage pools (pooled within 4 hours of transfusion) of up to 6 units of LR and nonleukoreduced (nLR) WBDP suspended in plasma; and
- Single units of LR and nLR WBDP suspended in plasma and tested within 4 hours before platelet transfusion as individual platelet units or as components of a post-storage pool.
The storage of platelet components at 20–24 °C [US Code of Federal Regulations Title 21 640.24(d)(1)] provides an environment for bacterial proliferation. There is no active hemovigilance in the US. Therefore, incidence rates of sepsis following platelet transfusion are estimates. From the FDA Issue Summary for the July 18, 2018 Blood Product Advisory Committee meeting (page 2): “Bacterial residual risk per transfused unit on the day of transfusion, despite primary culture, remains around 1/2,300, and fatal transfusion reactions from undetected contaminated platelet collections continue to occur. The reported rates of septic transfusion reactions from platelets vary from 1/100,000 by passive surveillance to 1/10,000 by active surveillance when testing with primary culture alone.”
FDA Final Guidance for mitigating the risk of transfusing bacterially contaminated platelets that could cause a septic reaction specifies the use of the Pan Genera Detection (PGD) Test as a permissible means of compliance with its recommendations (2). When PGD is used as a “safety measure” test, platelets may be stored for up to 7 days in storage containers cleared or approved for this purpose. At present such containers are available for leukocyte-reduced apheresis platelets stored in plasma and collected with sets from Terumo and Fresenius Kabi. There are two options in the Final Guidance for using the test as a “safety measure” to extend platelet storage for LRAP in plasma to 7 days: after storage day 5 following large volume delayed culture performed no sooner than 36 hours after collection or after storage day 4 following a primary culture performed no sooner than 24 hours after collection. In either case, the test needs to be performed only one time. In fact, a platelet that has outdated at the end of storage day 5 can be requalified and transfused on storage days 6 or 7 with a negative PGD Test.
Testing on the day of transfusion has been shown to detect bacterial contamination in platelets after they were distributed to hospitals as primary culture negative for bacteria (1). On the day of transfusion, bacteria may have entered logarithmic growth phase and proliferated to amounts that are expected to be higher than at 24 hours post-collection. Sampling and testing on day of transfusion adds a measure of safety by interdicting a proportion of highly contaminated units that pose a serious risk to transfusion recipients.
Like its predecessor, the Platelet PGD® Test, the Platelet PGDprime Test is a simple, rapid, day of transfusion test for the detection of bacterial contamination in platelets and is based on PGD® technology (1,3). The average cost of the device is USD$25.00. The test detects the presence of conserved antigens including lipoteichoic acid (LTA) and lipopolysaccharide (LPS) found on aerobic and anaerobic GP and GN bacteria, respectively. LTA and LPS targets are located on the surface of their respective bacteria and are primary constituents of the cell walls (4,5). LTA and LPS antigens are found on rapidly growing as well as stationary phase bacteria and their detection is possible by the use of specific antibodies (6,7). By combining the detection of LTA, LPS and other bacterial antigens in a single test device, it is possible to detect those species most frequently implicated in contaminated platelets (8,9).
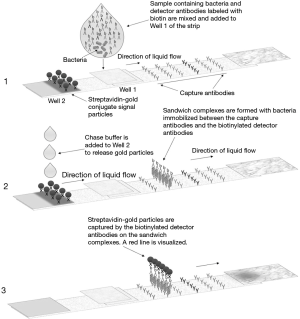
The Platelet PGDprime Test is a single-use, lateral flow, qualitative immunoassay comprising reagents, controls, disposables and a test device containing a test strip that is specific for the detection of aerobic and anaerobic GP and GN bacteria. Figure 1 illustrates the layout of the PGDprime Test. Samples from components noted above may be tested. Samples are mixed with the pretreatment reagent (Reagent 1A) and brought to the proper testing pH by the addition of the neutralizing detection reagent (Reagent 1B). The processed sample is transferred to Well 1 on the test device. As the sample migrates along the test strip, bacteria present in the sample bind to GP or GN bacteria-specific biotin-labeled detector antibodies thereby creating biotin-labeled sandwich complexes with capture antibodies immobilized on the nitrocellulose membrane. With addition of buffer reagent (Reagent 2) to Well 2 on the test device, streptavidin-coated 40-nm gold particles are released and flow across the nitrocellulose, “chasing” the processed sample. The streptavidin-coated gold binds to the biotin-labeled sandwich complexes that have formed in the presence of bacteria in the capture zones, creating a visible red/pink line in the zone(s) where bacteria have been captured. When the sample, detector antibodies and streptavidin gold have all reached the end of the strip, a control line becomes visible in the control window. When the background of the test result window has cleared of pink staining and the control line has formed, the test is valid and is interpreted by visual examination of the GP and GN test result window (1,3). The PGDprime Test has replaced the PGD Test.

Figure 2 Illustrates the sequential assay format of the PGDprime Test. Test sensitivity is driven by the large quantity of dissolved detector antibodies. PGDprime requires a sample size of only 150 µL. There are several procedural differences compared to the original PGD Test: no centrifugation, no humidity chamber, no temperature or humidity monitoring, no pellet resuspension, no precision pipetting, and no vigorous mixing except for nLR WBDP. The test can be performed with valid results at temperatures from 15–30 °C and is not susceptible to local ambient airflows owing to room and device ventilating fans. Each of the test reagents is added in six-drop increments with an error tolerance of ±2 drops. Only 2 minutes of exposure are required for sample pretreatment, although results are valid with pretreatment of up to 30 minutes. No loss of accuracy occurs when these conditions are present in multiple combinations (10).
Sensitivity
Limit of detection (LoD) (1,3)
The LoD of the original Platelet PGD Test was established for LRAP suspended in plasma and was determined for each of the ten organisms listed in Table 1. Testing was performed using 3 lots of the Platelet PGD Test with multiple operators and samples withdrawn from multiple LRAP (plasma) units and tested in replicates of 10. Dilution plate counting was used to assign a colony-forming unit (CFU)/mL concentration. The CFU/mL value of the sample when the Platelet PGD Test achieved 10/10 detection was defined as the assay’s LoD.
Table 1
| Organism | LoD CFU/mL |
|---|---|
| Bacillus cereus | 1.2×104 |
| Clostridium perfringens* (ATCC 13124) | 8.9×104 |
| Escherichia coli | 2.8×104 |
| Klebsiella aerogenes | 1.0×104 |
| Klebsiella pneumoniae | 2.0×104 |
| Pseudomonas aeruginosa | 8.2×103 |
| Serratia marcescens (ATCC 8100) | 8.6×105 |
| Staphylococcus aureus | 8.2×103 |
| Staphylococcus epidermidis | 9.2×103 |
| Streptococcus agalactiae | 5.5×104 |
*, anaerobe. Unless otherwise noted, bacterial strains were isolates from blood cultures or recovered from platelet contamination events. PGD, Pan Genera Detection; LRAP, leukocyte-reduced apheresis platelets; LoD, limit of detection; CFU, colony-forming unit.
The LoD of the PGDprime Test for each of these ten organisms was established by testing a three-level panel that was designed to bracket the LoD established for the PGD Test (11). Preparation of panel members to achieve precise CFU/mL concentrations is not possible. Each panel member was added to 10 LRAP (plasma) samples and tested using three lots of PGDprime Test and one lot of PGD Test, which served as a control. The PGDprime LoD was confirmed as the CFU/mL at which each lot achieved 100% detection, i.e., 10/10 for each lot. The LoD of the PGD Test was confirmed when 10/10 tests gave reactive results. Table 2 shows the organisms and CFU/mL concentrations at which the Platelet PGDprime Test and the Platelet PGD Test showed 100% detection.
Table 2
| Organism | PGDprime CFU/mL | Platelet PGD CFU/mL |
|---|---|---|
| Bacillus cereus | 2.7×104 | 2.7×104 |
| Clostridium perfringens* (ATCC 13124) | 2.4×105 | 2.4×105 |
| Escherichia coli | 5.6×104 | 5.6×104 |
| Klebsiella aerogenes | 3.3×104 | 3.3×104 |
| Klebsiella pneumoniae | 6.1×104 | 6.1×104 |
| Pseudomonas aeruginosa | 2.6×103 | 1.7×104 |
| Serratia marcescens (ATCC 43862) | 2.5×106 | 2.5×106 |
| Staphylococcus aureus | 2.1×103 | 1.8×104 |
| Staphylococcus epidermidis | 1.9×103 | 2.7×104 |
| Streptococcus agalactiae | 1.6×105 | 1.6×105 |
| Streptococcus oralis | 2.5×106 | ** |
*, anaerobe; **, non-reactive using the Platelet PGD Test. Unless otherwise noted, bacterial strains were isolates from blood cultures or recovered from platelet contamination events. PGD, Pan Genera Detection; LRAP, leukocyte-reduced apheresis platelets; CFU, colony-forming unit.
The LoD for Streptococcus oralis, which was not detectable by the original PGD, was estimated by preparing a bacterial stock, making serial dilutions in LRAP (plasma) and then performing dilution plate counts on the dilutions to assign CFU/mL values. PGDprime testing was performed on the dilutions to determine the lowest reactive dilution. This dilution was then further tested in three replicates in each of 3 lots of PGDprime. When all ten PGDprime results were reactive, the CFU/mL of the tested dilution was established as the LoD. The LoD for Streptococcus oralis was 1.95×106 CFU/mL. For additional testing, a Streptococcus oralis panel member targeted to fall within 0.5 Log of the LoD was prepared. The actual LoD would be between this concentration and the next higher dilution at 9.75×105 CFU/mL. Jacobs and colleagues have reported no morbidity associated with viridans group Streptococci at ≤5×106 CFU/mL (12).
Further testing was performed to assess and compare detection of the 11 organisms across platelet types: LRAP in plasma, LRAP in PAS-C/plasma, LR WBDP (PSPs), and nLR WBDP post-storage pools. Each mid-level panel member (LoD) was added to samples from 10 LR WBDP pools, 7 LRAP in PAS-C/plasma, 6 nLR WBDP pools and 9 LRAP in plasma, which served as control samples. Three lots of PGDprime Test were used. Detection was compared across platelet types. There was 100% detection across lots and platelet types at the LoD levels.
Analytical growth model studies for bacterial detection in platelets (1,3)
The Platelet PGDprime Test’s ability to detect bacteria growing in platelets was evaluated by determining the time to bacterial detection. Three lots of Platelet PGDprime were used in the study. All bacterial species and conditions were tested in duplicate with each lot. Platelet units were inoculated at the bacterial levels listed in Table 3. Neither Clostridium perfringens nor Streptococcus oralis grew reliably in LRAP (plasma) units, and therefore, were not included in this study.
Table 3
| Bacteria | Bacterial concentration at inoculation (CFU per bag) | Number of test samples detected by PGDprime at testing time point (n=6) | Second culture result | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 24 h | 36 h | 48 h | 60 h | 72 h | 84 h | 96 h | 108 h | |||
| Bacillus cereus (ATCC 7064) | 18.8 | 6 | 6 | – | – | – | – | – | – | Pos |
| Escherichia coli | 9.8 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 6 | Pos |
| Klebsiella aerogenes | 6.3 | 0 | 0 | 0 | 6 | 6 | – | – | – | Pos |
| 5.8 | 0 | 0 | 0 | 0 | 6 | 6 | – | – | Pos | |
| Klebsiella pneumoniae | 6.5 | 0 | 6 | 6 | – | – | – | – | – | Pos |
| Pseudomonas aeruginosa | 21 | 0 | 0 | 0 | 0 | 0 | 6 | 6 | – | Pos |
| Serratia marcescens (ATCC 43862) | 3.8 | 0 | 0 | 6 | 6 | – | – | – | – | Pos |
| Staphylococcus aureus (ATCC 27217) | 17.3 | 0 | 0 | 6 | 6 | – | – | – | – | Pos |
| Staphylococcus epidermidis (ATCC 49134) | 16.3 | 0 | 0 | 0 | 0 | 0 | 6 | 6 | – | Pos |
| Streptococcus agalactiae (ATCC 12927) | 13.8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6* | Pos |
*, reactive PGDprime results confirmed by subsequent testing at 120 h. Unless otherwise noted, bacterial strains were isolates from blood cultures or recovered from platelet contamination events. LRAP, leukocyte-reduced apheresis platelets; CFU, colony-forming unit.
Aliquots from platelet units in volumes of at least 50 mL were put into multiple, smaller platelet bags. At initiation (T=0), each platelet bag was inoculated with an estimated 10 CFU/mL of one bacterial species. One bag was inoculated with PBS only and held as a negative control for growth. After 2 hours, samples were withdrawn from inoculated bags for semi-quantitative culture. Inoculated bags with viable bacterial concentration between 1 and 30 CFU/mL were blind coded and continued in the study.
At 24-hour post-inoculation and every 12 hours thereafter each bag was sampled and tested in duplicate with each of 3 PGDprime Test lots until all PGDprime results were reactive at two consecutive time points. A second culture, including bacterial identification, was performed on each bag after reactive results were observed. The second culture confirmed the Platelet PGDprime Test results and the bacterial growth status of the bag. Time to detection was tabulated for each bacterial species. Study results are shown in Table 3. Not all bacterial species grew in one attempt. Two species, required additional growth attempts. Three growth attempts were required for Klebsiella aerogenes. Streptococcus agalactiae did not grow in four attempts but did grow successfully with a fifth attempt.
Ultra-low inoculum study (1,3)
The objective of this study was to demonstrate that the Platelet PGDprime Test was able to detect bacteria missed by early culture due to sampling error. This study was performed using three lots of the Platelet PGDprime Test. LRAP in plasma, LRAP in PAS-C and plasma, and pools of LR WBDP were inoculated with three bacterial species: a GP (Bacillus cereus), a GN (Klebsiella pneumoniae) and a slower growing organism (Staphylococcus epidermidis). A negative control was also prepared by inoculating PBS into the same platelet matrices. Inoculated units were blind coded so that the technologist performing testing was unaware of the expected results.
Bacteria were inoculated at very low titer (targeting <200 CFU per bag) into each bag, mixed for 1 to 2 hours and then sampled for initial culture testing. Ten 8 mL samples were removed from each bag. A 1 mL volume from each sampling was added to each of four agar plates and incubated under aerobic conditions only, as no anaerobic organisms were inoculated. Plates were monitored for growth. An inoculated bag was excluded from further study if colonies were observed on 10 of the 10 samples (indicating no culture sampling error). If colonies were observed on fewer than 10 of the 10 samples (indicating culture sampling error), the inoculated bag qualified for study inclusion.
Samples collected at 24-hours post-inoculation and every 12 hours thereafter were tested in duplicate using three PGDprime Test lots. For LR WBDP, one volume of inoculated platelet was combined with five volumes of uninoculated pooled platelets to prepare a pooled sample at the time of testing. Testing continued until reactive results were observed on all six PGDprime Test devices for at least two consecutive samplings. A second culture, including bacterial identification, was performed on each bag after reactive results were observed. The second culture confirmed the platelet PGDprime Test results and the bacterial growth status of the bag.
Ultra-low inoculum study—LRAP suspended in plasma (1,3)
Of ten bags inoculated, five supported bacterial growth (Table 4). Of 50 initial culture samples taken from these five bags, 35 demonstrated sampling error resulting in false negative culture results. PGDprime Test results for these five bags were reactive starting 24 hours after inoculation of the bag. The times to detection of the two bags inoculated with Bacillus cereus were 24 and 36 hours. Time to detection for the bag inoculated with Klebsiella pneumoniae was 36 hours and was 96 hours for the two bags inoculated with Staphylococcus epidermidis. The Platelet PGDprime Test was able to detect bacterial contamination in LRAP in plasma units when an early culture did not reliably detect bacteria due to sampling error.
Table 4
| Bacteria | Bacterial concentration at inoculation (CFU/bag) | Initial culture samples positive | Number of test samples detected by PGDprime at testing time point (n=6) | Second culture result | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 24 h | 36 h | 48 h | 60 h | 72 h | 84 h | 96 h | 108 h | ||||
| Bacillus cereus (ATCC 7064) | |||||||||||
| Bag 1 | 117.3 | 5 of 10 | 6 | 6 | – | – | – | – | – | – | Pos |
| Bag 2 | 11.73 | 3 of 10 | 0 | 6 | 6 | – | – | – | – | – | Pos |
| Klebsiella pneumoniae | |||||||||||
| Bag 3 | 1.45 | 0 of 10 | 0 | 6 | 6 | – | – | – | – | – | Pos |
| Staphylococcus epidermidis (ATCC 49134) | |||||||||||
| Bag 4 | 17.0 | 0 of 10 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 6 | Pos |
| Bag 5 | 162.8 | 7 of 10 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 6 | Pos |
LRAP, leukocyte-reduced apheresis platelets; CFU, colony-forming unit.
Ultra-low inoculum study—LRAP suspended in PAS-C and plasma (1,3)
Of ten bags inoculated, four supported bacterial growth (Table 5). Of 40 initial culture samples taken from these four bags, 16 demonstrated sampling error resulting in false negative culture results. PGDprime Test results for these four bags were reactive starting at 36 hours after inoculation of the bag. The time to detection of the bag inoculated with Bacillus cereus was 36 hours. Time to detection for the two bags inoculated with Klebsiella pneumoniae was 36 hours. For the bag inoculated with Staphylococcus epidermidis the time to detection was 96 hours.
Table 5
| Bacteria | Bacterial concentration at inoculation (CFU/bag) | Initial culture samples positive | Number of test samples detected by PGDprime at testing time point (n=6) | Second culture result | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 24 h | 36 h | 48 h | 60 h | 72 h | 84 h | 96 h | ||||
| Bacillus cereus (ATCC 7064) | ||||||||||
| Bag 1 | 56.1 | 6 of 10 | 0 | 6 | 6 | – | – | – | – | Pos |
| Klebsiella pneumoniae | ||||||||||
| Bag 2 | 25.1 | 5 of 10 | 0 | 6 | 6 | – | – | – | – | Pos |
| Bag 3 | 188.4 | 9 of 10 | 0 | 6 | 6 | – | – | – | – | Pos |
| Staphylococcus epidermidis (ATCC 49134) | ||||||||||
| Bag 4 | 66.1 | 4 of 10 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | Pos |
LRAP, leukocyte-reduced apheresis platelets; PAS, platelet additive solution; CFU, colony-forming unit.
The Platelet PGDprime Test was able to detect bacterial contamination in LRAP suspended in PAS-C and plasma when early culture did not reliably detect bacteria due to sampling error.
Ultra-low inoculum study—PSPs of LR WBDP suspended in plasma
Of ten bags inoculated, four supported bacterial growth (Table 6). Of 40 initial culture samples taken from these four bags, 29 demonstrated sampling error resulting in false negative culture results. Prior to testing, samples were prepared by combining one volume of inoculated platelet with five volumes of uninoculated pooled platelets. PGDprime Test results for these four bags were reactive starting at 36 hours after inoculation of the bag. Time to detection of both bags inoculated with Bacillus cereus was 36 hours. The time to detection of Klebsiella pneumoniae was 36 hours, while time to detection of Staphylococcus epidermidis was 96 hours. The Platelet PGDprime Test was able to detect bacterial contamination in PSPs of LR WBDP suspended in plasma when early culture did not reliably detect bacteria due to sampling error.
Table 6
| Bacteria | Bacterial concentration at inoculation (CFU/bag) | Initial culture samples positive | Number of test samples detected by PGDprime at testing time point (n=6) | Second culture result | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 24 h | 36 h | 48 h | 60 h | 72 h | 84 h | 96 h | ||||
| Bacillus cereus (ATCC 7064) | ||||||||||
| Bag 1 | 8.1 | 1 of 10 | 0 | 6 | 6 | 6 | – | – | – | Pos |
| Bag 2 | 75.4 | 9 of 10 | 0 | 6 | 6 | 6 | – | – | – | Pos |
| Klebsiella pneumoniae | ||||||||||
| Bag 3 | 24.8 | 1 of 10 | 0 | 0 | 6 | 6 | 6 | 6 | 12† | Pos |
| Staphylococcus epidermidis (ATCC 49134) | ||||||||||
| Bag 4 | 65.5 | 0 of 10 | 0 | 0 | 0 | 0 | 0 | 0 | 12† | Pos |
†, Operator ran a second bag sample on a second set of devices to confirm reactivity status at last time point. All results were consistently reactive. LR, leukoreduced; WBDP, whole blood-derived platelets; CFU, colony-forming unit.
Potentially interfering substances (1,3,13)
This study evaluated substances or sample conditions that might interfere with the ability of the Platelet PGDprime Test to correctly identify bacterial-negative platelet samples as non-reactive (NR) and bacteria-positive samples as reactive. The substances and conditions are listed in Table 7. All testing was performed using a 12-member panel comprising 11 bacteria-positive and 1 bacteria-negative members. Three lots of the Platelet PGDprime Test were used. LRAP in plasma and LR WBDP/LR WBDP pooled and nLR WBDP/nLR WBDP pooled were tested. At least five examples of each potentially interfering condition were tested for each platelet type, with the exception of rheumatoid factor (RF) and human anti-mouse antibody (HAMA), where ten samples were tested.
Table 7
| Source of interference | Substance tested | Substance level |
|---|---|---|
| Donor conditions | Autoimmune antibodies | Ds-DNA (10–360 IU/mL) |
| ANA (positive, qualitative test) | ||
| RF: 16.9–272 IU/mL | ||
| Human anti-mouse antibody (HAMA): 10.5–182 ng/mL | ||
| Hypergammaglobulinemia | IgA (505–744 mg/dL) | |
| IgG (2,105–3,901 mg/dL) | ||
| IgM (461–933 mg/dL) | ||
| Lipemia | 305–553 mg/dL | |
| Hypercholesterolemia | 352–1,230 mg/dL | |
| Hyperproteinemia | ≥9 g/dL | |
| Hypoproteinemia | 2.84–3.98 g/dL | |
| Sample conditions | Hemolysis | 0–350 µg/dL |
| pH | 5.5–8.5 | |
| Platelet concentration (% normal/native) | 50–200% average concentration | |
| Red blood cells (concentration in %) | 0–0.7% | |
| White blood cells | 4×104–4×105 cells/mL for WBDP | |
| Platelet additive solution | 0 and 100% for LRAP |
ANA, antinuclear antibody; RF, rheumatoid factor; LRAP, leukocyte-reduced apheresis platelets; WBDP, whole blood-derived platelets.
With the exception of one elevated IgM sample, there were no effects of the substances/conditions tested on the performance of the Platelet PGDprime Test when testing bacteria-positive panel members. One elevated IgM sample yielded a repeatable false negative result with Escherichia coli, while all other bacteria were detected in the presence of this IgM sample. A possible explanation for this result is that the sample contains a high titer of antibodies specific to Escherichia coli.
The potential interferents purchased for use in evaluating donor conditions were not provided as sterile materials from the supplier. Six HAMA samples and one antinuclear antibody (ANA) sample produced reactive results on single GN lines, results that are consistent with the presence of bacterial antigens (the only mouse antibody used in the test that could potentially be interfered with detects GP bacteria, not GN species). One ds-DNA sample and two hyperproteinemia samples produced signals on multiple capture lines, indicative of non-specific reactions. All other samples representing donor conditions and all samples representing sample conditions produced NR results when tested in platelet samples absent bacterial panel members. One elevated IgM, one elevated IgG and four hyperproteinemia samples (>10 g/dL) did not flow in the assay yielding invalid test results.
Reproducibility of detection (1,3)
During the LoD Study, 1,356 PGDprime test results were generated using 3 lots of PGDprime Test, 11 bacterial species and 4 platelet types. The Platelet PGDprime Test detected bacteria accurately and reproducibly in each of the lots and platelet types.
Specificity
Specificity of the Platelet PGDprime Test was assessed for LRAP suspended in plasma; LRAP suspended in PAS-C and plasma; LR-WBDP and nLR WBDP, some of which were also combined and tested as LR-WBDP pooled and nLR WBDP pooled; and PSP. Age of platelets tested in the study ranged from 2 to 6 days post-collection. The specificity study was performed at multiple sites (14). Three lots of PGDprime were used and lots were evenly distributed across sites.
To be included in the specificity study, each platelet sample had to have both a PGDprime result and a negative culture result by traditional agar plate culture (APC). Bacterial status of each unit was determined by APC in a blinded study independent of PGDprime testing. If no colonies were observed under either aerobic or anaerobic conditions after 3–7 days, the unit was deemed to be bacteria-negative.
Repeat testing was performed on samples with initially invalid (INV) or initially reactive (IR) test results. The study protocol specified performing one repeat test for any sample with an INV result and required two repeat tests for a sample with an IR result (GP or GN or both). Interpretations of PGDprime results were (using only valid tests):
- Repeatedly reactive (RR) if the initial and at least one of the two repeat PGDprime tests were reactive. This was considered a positive result;
- NR if the initial PGDprime test was NR or if the initial PGDprime test was reactive but both repeat PGDprime tests were non-reactive.
Specificity was calculated for each platelet type as the percent of culture-negative units that would lead to a PGDprime-NR interpretation. One-sided 95% confidence limits were calculated using the Wilson method. Table 8 summarizes the specificity observed for PGDprime with various platelet types. Out of 3,800 unique samples, 5 were IR, 4 were NR upon retest. One sample was classified as indeterminate as only one repeat test was performed by the test site (this single repeat was negative). The overall test specificity was determined to be 100% [lower 1-sided 95% confidence limit (LCL) 99.9%]. No samples were RR.
Table 8
| Unit type | Results | All | Initial specificity | Final specificity | |||||
|---|---|---|---|---|---|---|---|---|---|
| IR | NR | IND | Estimate | Lower 1-sided 95% CL | Estimate | Lower 1-sided 95% CL | |||
| LRAP | 1 | 1,598 | 0 | 1,599 | 99.9% | 99.7% | 100.0% | 99.8% | |
| PAS | 2 | 295 | 0 | 297 | 99.3% | 98.0% | 100.0% | 99.1% | |
| LR-WBDP | 0 | 611 | 0 | 611 | 100.0% | 99.6% | 100.0% | 99.6% | |
| LR-WBDPp | 0 | 75 | 0 | 75 | 100.0% | 96.5% | 100.0% | 96.5% | |
| nLR WBDP* | 1 | 501 | 1 | 503 | 99.6% | 98.8% | 99.8% | 99.1% | |
| 502 | 100.0% | 99.5% | |||||||
| nLR WBDPp | 1 | 64 | 0 | 65 | 98.5% | 93.4% | 100.0% | 96.0% | |
| PSP | 0 | 650 | 0 | 650 | 100.0% | 99.6% | 100.0% | 99.6% | |
| All* | 5 | 3,794 | 1 | 3,800 | 99.8% | 99.7% | 100.0% | 99.9% | |
| 100.0% | 99.9% | ||||||||
*, reported both with (N=503) and without (N=502) the sample classified as IND because the IR for that sample was only followed up by a single additional test making it impossible to interpret as either NR or RR. IR, initially reactive; NR, non-reactive; RR, repeatedly reactive; IND, indeterminate; CL, confidence level; LRAP, leukocyte-reduced apheresis platelets; LR, leukoreduced; PAS, platelet additive solution; WBDP, whole blood-derived platelets; WBDPp, whole blood-derived platelets pooled; nLR, nonleukoreduced; PSP, pre-storage pools.
Human anti-animal (heterophile) antibodies may be present in human plasma. In the first-generation PGD Test, these reacted with the Fc portions of both the capture antibody and the detector antibody, thereby creating a sandwich complex resulting in a false-positive result. This heterophile reactivity was responsible for most of the observed 0.5% false-positive rate. This problem has been corrected by the removal of the Fc fragment from the detector antibodies (15).
To prevent the development of these unwanted sandwich complexes, PGDprime uses F(ab')2 fragments for 6 of the 7 detector antibodies. F(ab')2 are fragments created from the enzymatic cleavage of the Fc portion of the whole antibody. With the Fc region removed from the detector antibody, false-positive sandwich complexes cannot be created. For the seventh detector antibody, an added animal immunoglobulin is used to block these interactions. As noted above, the specificity of the PGDprime Test 100%. (LCL 99.9%) (1).
Recent developments
In 2018, four transmissions of Acinetobacter calcoaceticus-baumannii complex by platelet transfusion were reported. These resulted in transfusion recipient morbidity and mortality (16). In two of the morbidity cases, the PGD Test had been used as a safety measure. Neither PGD nor PGDprime had been designed to efficiently detect Acinetobacter spp. since these had not been reported as platelet contaminants. An updated version of PGDprime with improved Acinetobacter detection was developed and optimized and has been validated in laboratory studies (17).
Hospital experience
Many sites have published reports of culture-confirmed true positive PGD Test results in primary culture-negative platelets (18-28). This real-world experience documents the effectiveness of the test in interdicting bacterially contaminated components that have been presumed to be safe by a negative primary culture result. For example, Jacobs et al. reported nine such results from multiple hospitals (20) and Mintz et al. reported 36 also from multiple sites (27). More than 1.4 million PGD Tests have been distributed to hospitals in the US with no fatal septic reaction reported.
Conclusions
The PGDprime Test is a sensitive, specific, easy to use day of transfusion device that has been extensively validated in laboratory testing and clinical use. The test has effectively interdicted the transfusion of many bacterially contaminated platelets. More than 1.4 million platelets have been distributed to users with no reported fatal septic transfusion reactions with tested platelets. It is FDA cleared to test all licensed platelets in the US except those that have been pathogen-reduced. It is specifically cleared as a “safety measure” to extend the outdate of LRAP in plasma to 7 days in bags cleared or approved for this purpose, thereby significantly improving availability while reducing expenses associated with platelet outdates. As a result of reduced outdating, extending platelet dating to 7 days has been reported to save more money than the test costs in many blood centers and hospital transfusion services (29).
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Sandra Ramirez-Arcos) for the series “Bacterial Contamination of Platelet Components” published in Annals of Blood. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aob-2020-bcpc-01). The series “Bacterial Contamination of Platelet Components” was commissioned by the editorial office without any funding or sponsorship.Dr. PDM and Dr. RPV are employees of Verax Biomedical, Inc. Dr. RPV discloses he holds a pending patent for a sequential lateral flow device. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Platelet PGDprime® Test. Available online: https://www.veraxbiomedical.com/wp-content/uploads/2020/02/US-Platelet-PGDprime-Test-Insert.1.pdf
- Bacterial Risk Control Strategies for Blood Collection Establishments and Transfusion Services to Enhance the Safety and Availability of Platelets for Transfusion. Guidance for Industry. Silver Spring, MD: U.S. Department of Health and Human Services, Food and Drug Administration, Center for Biologics Evaluation and Research. September 2019, updated December 2020. Available online: https://www.fda.gov/media/123448/download
- Vallejo RP, Shinefeld L, LaVerda D, et al. Performance profile of an updated safety measure rapid assay for bacteria in platelets. Transfusion 2020;60:2622-32. [Crossref] [PubMed]
- Fischer W. Physiology of lipoteichoic acids in bacteria. Adv Microb Physiol 1988;29:233-302. [Crossref] [PubMed]
- Rietschel ET, Brade H, Holst O, et al. Bacterial endotoxin: Chemical constitution, biological recognition, host response, and immunological detoxification. Curr Top Microbiol Immunol 1996;216:39-81. [Crossref] [PubMed]
- Jackson DE, Wong W, Largen MT, et al. Antibodies to immunodeterminants of lipoteichoic Acids. Infect Immun 1984;43:800-803. [Crossref] [PubMed]
- Lugowski C, Niedziela T, Jachymek W. Anti-endotoxin antibodies directed against Escherichia coli R-1 oligosaccharide core-tetanus toxoid conjugate bind to smooth, live bacteria and smooth lipopolysaccharides and attenuate their tumor necrosis factor stimulating activity. FEMS Immunol Med Microbiol 1996;16:31-8. [Crossref] [PubMed]
- Kuehnert MJ, Roth VR, Haley NR, et al. Transfusion-transmitted bacterial infection in the United States, 1998 through 2000. Transfusion 2001;41:1493-9. [Crossref] [PubMed]
- Yomtovian R. Bacterial contamination of blood: lessons from the past and road map for the future. Transfusion 2004;44:450-60. [Crossref] [PubMed]
- Shinefeld L, Best N, Williams M, et al. User guardband study to determine the robustness of the Verax platelet PGD
prime test for bacteria in platelets. Transfusion 2018;58:228A. - Shinefeld L, LaVerda D, Best N, et al. Validation of the analytical sensitivity of commercial scale lots of the verax PGD
prime test for bacteria in platelets. Transfusion 2019;59:155A. - Jacobs MR, Good CE, Lazarus HM, et al. Relationship between bacterial load, species virulence, and transfusion reaction with transfusion of bacterially contaminated platelets. Clin Infect Dis 2008;46:1214-20. [Crossref] [PubMed]
- Shinefeld L, Best N, Williams M, et al. Robustness of the Verax PGD
prime test for bacteria in platelets to interfering substances in plasma. Transfusion 2018;58:239A-240A. - Shinefeld L, Hornbaker N, Rasmusson P, et al. Validation of the specificity of the PGD
prime test for bacteria in platelets with commercial scale lots. Transfusion 2019;59:40A. - Best N, Lamkin M, Shinefied L. Reducing false‐positive reactions in a rapid test for bacteria in platelets. Transfusion 2015;55:193A.
- Jones SA, Jones JM, Leung V, et al. Sepsis Attributed to Bacterial Contamination of Platelets Associated with a Potential Common Source - Multiple States, 2018. MMWR Morb Mortal Wkly Rep 2019;68:519-23. [Crossref] [PubMed]
- LaVerda D, Shinefeld L, Best N, et al. Updating the Platelet PGD
prime Rapid Test® for bacteria in platelets to detect acinetobacter. Transfusion 2020;60:73A-74A. - Jacobs MR, Good CE. Residual Bacterial Contamination of Apheresis Platelets Following Early Culture – Results of a Multi-Site Study of 18,449 Units Using the Verax Pan Genera Detection Assay. Transfusion 2010;50:30A.
- Jacobs MR, Smith D, Heaton WA, et al. Detection of Bacterial Contamination in Prestorage Culture Negative Apheresis Platelets on Day of Issue with the PGD Test. Transfusion 2011;51:199A. [Crossref]
- Jacobs MR, Smith D, Heaton WA, et al. Detection of bacterial contamination in prestorage culture-negative apheresis platelets on day of issue with the Pan Genera Detection test. Transfusion 2011;51:2573-82. [Crossref] [PubMed]
- Harm SK, Delaney M, Charapata M, et al. Use of a Time-of-Issue Bacteria Screening Test for Whole Blood Derived Platelets at Two Centralized Transfusion Services. Transfusion 2012;52:13A.
- Korte LG, Sowell J G, Bracey A. Implementation of PGD Testing for Whole Blood Derived Platelets. Transfusion 2012;52:45A.
- Hornbaker N, Rasmusson P, Lee W, et al. Pan Genera Detection (PGD®) Testing for Bacteria in Platelets at 50 US Institutions. Transfusion 2012;52:206A.
- Alrabeh R, Sowell J, Korte LG, et al. Verax Pan Genera Detection (PGD) Test for Platelet Screening: A 5-year Retrospective Analysis in a High-volume Transfusion Service. Transfusion 2016;56:72A.
- Frye M, van Antwerpen K, Stevenson ME, et al. Improved Platelet Stewardship by Rapid Bacterial Testing at a Blood Center. Transfusion 2019;59:151A-152A.
- Alrabeh R, Korte L, Reyes M, et al. Long-term Experience with Rapid Screening for Platelet Bacterial Contamination in a High-volume Transfusion Service. Ann Clin Lab Sci 2019;49:748-55. [PubMed]
- Mintz PD, Sanders J, Blair J, et al. Confirmed Positive Bacterial Detection in Platelet Concentrates by a Rapid Test after Negative Primary Culture. Transfusion 2019;59:66A.
- Metzler G, Music-Aplenc L. Prevention of the Transfusion of Bacterially Contaminated Platelets at a Children's Hospital with the Platelet PGD Test. Transfusion 2020;60:247A-248A.
- Mintz PD, Sanders JR. Outdate reduction and cost savings with rapid testing for seven‐day platelet storage. Ann Clin Lab Sci 2020;50:404-7. [PubMed]
Cite this article as: Mintz PD, Vallejo RP. The PGDprime immunoassay for detection of bacterial contamination in platelet concentrates: sensitivity and specificity. Ann Blood 2021;6:19.


