
FDA guidance on bacterial contamination risk control strategies to enhance the safety and availability of platelets: advantages and limitations
Introduction
Bacterial contamination of platelet products has been a major challenge due to storage of these products at room temperature to preserve platelet function. Initial small bacterial loads can increase to very high inocula during room temperature storage of products, with transfusion reactions generally only occurring with bacterial loads above 105 CFU/mL (1). The problems of bacterial contamination, recognized soon after the introduction of platelet products into clinical use (2), as well as platelet function have limited storage to a maximum of 7 days after collection, with shelf-life usually limited to 5 days.
In the US, the production, use and shelf-life of platelets is set by the Center for Biologics Evaluation and Research (CBER) of the US Food and Drug Administration (FDA), which will be referred to as “FDA” throughout this review, based on multiple FDA regulations issued in Title 21 of the Code of Federal Regulations. In 1984 the shelf life of platelets was increased from 5 to 7 days by FDA, but a subsequent increase in reports of septic transfusion reactions led to storage time being reduced back to 5 days in 1986 (3). While there was no FDA-mandated regulatory requirement to institute measures to decrease bacterial contamination at that time, two US accrediting agencies, the College of American Pathologists (CAP) and AABB, subsequently introduced standards to reduce bacterial contamination in 2002 and 2004, respectively. The action by CAP in 2002 was to introduce a Phase I (optional) Laboratory Accreditation Checklist recommendation “Does the laboratory have a system to detect the presence of bacteria in platelet components?” This became a Phase II (mandatory) requirement in 2004. The AABB Blood Bank/Transfusion Service Standards Program Unit approved standard 5.1.5.1 mandated testing for bacteria in all platelet components in March 2003 for implementation by March 2004. This stated that “The blood bank or transfusion service shall have methods to limit and detect bacterial contamination in all platelet components.” This AABB standard was generally implemented by culture of aliquots of apheresis collections 24 hours after collection at collection centers, and by pH or glucose level testing of pooled, whole blood derived (WBD) units by hospital transfusion services at time of issue (4). Introduction of prepooling of WBD units at collection centers from 2007 allowed these products to be cultured as well (5). These cultures, now referred to as “primary cultures,” were used by all US platelet producing facilities from 2004, initially by culture of 3–4 mL of products, usually apheresis platelet collections before being split into individual doses, in BacT/ALERT blood product aerobic bottles or in BDS (later eBDS) culture pouches (4,6). In 2007 many suppliers subsequently increased the volume of platelets cultured in BacT/ALERT aerobic bottles from 4 mL to 8 mL per bottle (7).
Bacterial contamination of platelet products has been documented in many countries, and a recent meta-analysis of contamination rates detected by primary culture identified 22 studies published between 2003 and 2018 with suitable data; 21 were of apheresis collections (just over 4 million), 4 were WBD pools (approx. 140,000), and 15 were buffy coat pools (1.47 million) (8). The mean contamination rate per million components were as follows: overall, for all products it was 510 (95% CI: 380–670; 1 in 1,961); the mean was lower for apheresis platelets (230, 95% CI: 180–280) and WBD pools (380, 95% CI: 150–700), and higher for buffy coat pools (1,120, 95% CI: 510–1,960). This study concluded that larger sample volumes increased sensitivity, while bacterial contamination rates decreased over time.
The National Blood Collection and Utilization Survey (NBCUS) performs annual surveys of all US blood collection centers, all hospitals performing at least 1,000 surgeries annually, and a 40% random sample of hospitals performing 100 to 999 surgeries annually. This data is used to generate national estimates of units of blood and components collected, deferred, distributed, transfused, and outdated. The 2017 NBCUS, the most recent survey available, showed that 2.56 million platelet units were collected and distributed (9-11). Of these, 1.93 million units (75%) were transfused in the form of 1.85 million apheresis units (95.8%), with 68,000 being pathogen reduced units (3.7% of apheresis units), and 82,000 WBD pools (4.2%). Secondary testing was reported on 114,000 platelet units (6.0% of units transfused, excluding pathogen reduced units), 51,000 by culture (yielding 13 confirmed positives; 255 per million, 95% CI: 150–436) and 63,000 by rapid test (yielding 10 confirmed positives; 159 per million, 95% CI: 87–292). These contamination rates are comparable to that found by active surveillance (by culture of all platelet units at time of issue) of 348 per million (95% CI: 250–487) (12), with the rate detected by rapid test being lower due to its higher bacterial load detection limit compared to culture, but comparably clinically relevant as low bacterial loads do not result in septic reactions (1). A total of 37 bacterial infections were reported by NBCUS from these transfusions, a rate of 2.3 (95% CI: 1.3–3.4) per million transfusions. This incidence of septic reactions is considerably lower than that reported (19.5 per million) for the years 2010–2016 by health care facilities participating in the National Healthcare Safety Network Hemovigilance Module (13), as well as the incidence detected by active surveillance (by culture of all platelet units at time of issue) of 82 per million for the years 2007–2017 (12). These major differences in septic reaction rates highlight the problem of recognition and reporting these reactions and using these rates to assess the effect of various interventions, such as changes in timing of testing and volume of platelets cultured in primary culture protocols; this is acknowledged in the FDA guidance, which acknowledges variations in reports of these reactions ranging from 10 to 100 per million transfusions (14,15). This is a key point that will be referred to later in discussion of various enhanced primary culture strategies.
Extension of the shelf-life of platelet products to 7 days has long been a goal to improve the availability of these products. An early program to extend apheresis platelet shelf life to 7 days was instituted in 52 US centers from September 2005 through April 2008 (16). This study, named the PASSPORT study, was sponsored by CaridianBCT, Inc. and Fenwal, Inc. using bags approved for 7-day storage, and study requirements and evaluation criteria were developed with the FDA. Primary culture of 388,903 apheresis collections was performed 24 h after collection using 8–10 mL samples split into aerobic and anaerobic bottles. Expired units were cultured again (secondary culture) using the same culture protocol to assess efficacy. Primary culture yielded 90 contaminated collections (231 per million, comparable to the mean of 230 per million reported in a meta-analysis study) (8), while four of 6,039 units (662 per million) were culture positive at outdate, a rate comparable to other studies (12). The study was deemed a failure as it did not meet the criteria set (primarily that the outdate contamination rate would be lower than the primary culture rate), and platelet outdate continued to be 5 days and the higher primary culture volume was deemed a failure and not introduced into routine use.
More recent developments have been regulatory approval for two methods for extending outdate to 7-days using a secondary test, Platelet PGDprime Test (Verax Biomedical), performed on 150 µL samples within 24-hour of use, and culture of 8–10 mL per bottle using BacT/ALERT aerobic and anaerobic culture bottles (bioMérieux) on days 4–6 (17,18). BACTEC Platelet Aerobic/F and Anaerobic/F Culture vials (Beckton Dickinson) have recently been approved for platelet testing, but their use is currently limited to quality control testing of platelet products as primary testing (19). The VersaTREK system (ThermoFisher) has also been shown to detect bacterial contamination of pooled buffy coat platelets (20). Current strategies for limiting bacterial contamination of platelet products under regulatory requirements and standards of care are shown in Table 1.
Table 1
| Strategy | Primary culture minimum volume per bottle (bottle type) | Time after collection (hours) | Incubation time before release (hours) | Shelf life (days) |
|---|---|---|---|---|
| Primary culture of apheresis collections and pre-storage WBD pools | 8 mL (aerobic) per collection or pool | ≥24 | ~12 | 5 |
| Secondary testing of apheresis platelet units in plasma with FDA cleared Safety Measure rapid test within 24 hours of transfusion | NA | NA | NA | Up to 7 |
| Secondary culture of apheresis units on day 3 | 5 mL (aerobic) per unit | NA | NA | 5 |
| Primary rapid test on single WBD units or post storage WBD pools within 4 hours of transfusion | NA | NA | NA | 5 |
| Pathogen reduction of apheresis units performed within 24 h of collection | NA | ≤24 | NA | 5 |
*, information from (15). Guidance specifies that times in hours are defined as exact time after collection or sampling, while times in days are defined as any time on day specified.
In 2014, FDA published draft guidelines for adoption of certain newer strategies to control the risk of bacterial contamination in 5-day platelets and to extend platelet dating up to 7 days (14); these draft guidelines were updated in 2016, and three FDA Blood Products Advisory Committee Meetings have been held to discuss these guidelines. At the July 2018 meeting the Committee was asked to address several strategies proposed by FDA to reduce bacterial contamination of platelet products (21). These strategies included primary testing only using increased culture volumes and/or delayed sampling, primary and secondary testing methods, and use of pathogen reduction technology. These strategies will be discussed and pertinent literature reviewed, with critical analysis of the quality of studies and data sources regarding bacterial contamination and septic transfusion reactions.
While the above discussion shows that there has been considerable concern about bacterial contamination for many decades, no major change other than pathogen reduction has occurred in the US to further mitigate this problem since the introduction of primary culture in 2004. The FDA guidance document acknowledges the poor sensitivity of primary culture (<50%) and has the stated goal of providing guidance on measures to improve the safely of platelet products (15). Fatalities from transfusion of bacterially contaminated platelets continue to occur, with several recent cases being of major concern as contaminants included the very virulent Gram negative pathogens, Klebsiella pneumoniae and Acinetobacter calcoaceticus-baumannii complex, as well as the Gram positive anaerobe, Clostridium perfringens (22-25). Of particular concern are reports of four units contaminated with Acinetobacter calcoaceticus-baumannii complex occurring in California, Utah, Massachusetts and North Carolina, with two of the units being pathogen reduced, as well as the fact that all these isolates are genetically related and appear to have a common source (22-25).
Assessment of the incidence of bacterial contamination of platelet products
The first issue in assessing the value of mitigating strategies is to obtain information on the incidence of bacterial contamination of the various platelet products at time of transfusion or outdate, ideally before and after introducing a mitigating strategy. This is readily and inexpensively achieved by testing for the presence of bacteria by quantitative or qualitative culture, or by testing for bacterial products by detection of bacterial cell wall or nucleic acid components (26-29). Bacterial contamination can also be assessed from reports of septic transfusion reactions, which is of much more limited value as discussed earlier. These factors have been studied at the author’s institution for almost three decades by culture of platelet units at time of issue and evaluation of transfusion reactions (1,12,26,30-39). These studies have documented the following. (I) Bacterial contamination continues to the present time, despite introduction of measures such as improved skin disinfection protocols, diversion pouches, and primary culture, showing the need for additional measures. (II) Active bacterial surveillance by culture of platelets at time of issue is the key to understanding the extent of the problem and the effect of interventions. (III) Primary culture was effective in removing many of the fastest-growing, most virulent bacterial species, but not in reducing the contamination rate or eliminating septic reactions and fatalities. (IV) Recognition and especially reporting of septic reactions are poor, so assessment of the value of interventions based on septic reaction reports is of limited value. (V) The clinical features of septic reactions changed after introduction of primary culture, frequently being delayed, less severe and more difficult to differentiate from other transfusion reactions. (VI) Prepooling and primary culture of WBD platelets has been very beneficial, reducing contamination rate to the range associated with apheresis platelets. (VII) In contrast, contamination rate of apheresis platelets did not change after introduction of primary culture and was not affected by primary culture method or volume of platelets cultured.
Septic transfusion reaction surveillance
While septic and fatal transfusion reaction reports are valuable in assessing the safety of platelet products, they have many limitations (34,40-43). Transfusion reaction surveillance programs can be active or passive. In active clinical surveillance programs, dedicated hemovigilance officers monitor transfused patients and check that appropriate investigation and reporting is performed or has occurred. Another form of active surveillance is the at-issue culture program referred to above, which detects all bacterially contaminated units and allows correlation of bacterial species and load with transfusion reactions and primary culture methods (1). In contrast, passive surveillance, the predominant surveillance method, relies on caregivers recognizing, investigating and reporting transfusion reactions consistent with septic reactions. These factors are demonstrated in a study from the author’s institution, which showed failure of passive surveillance to detect septic reactions and lack of specificity of septic reaction criteria (12,36). As noted earlier, the introduction of primary culture changed the severity, but not the incidence, of bacterial contamination and septic transfusion reactions associated with apheresis platelets, but did decrease the incidence considerably with pooled whole-blood derived units (37).
In long-term studies at two US institutions with active surveillance programs, Johns Hopkins Hospital and Medical Institutions (clinical surveillance) and the author’s institution (bacteriological surveillance), the incidence of septic reactions to apheresis and prepooled platelets released as negative after primary culture has been remarkably constant over time, ranging from 36 to 50 cases per million transfusions (1,26,27,36,37,44,45). In contrast, the incidence was 5- to 10-fold lower, ranging from 1 to 10 cases per million transfusions, with passive surveillance systems (7,46-48). The limitations of passive surveillance are illustrated by the strict criteria used to diagnose septic transfusion reactions in the UK. This requires that the same bacterial species be isolated from the platelet unit and the patient’s blood culture; investigation of 856 reported cases of suspected platelet transfusion associate sepsis from 2012 through 2019 by the UK Serious Hazards of Transfusion (SHOT) group resulted in only one case determined to be a septic reaction (43). The SHOT report noted “Haemovigilance systems for bacterial TTI (transfusion transmitted infection) are passive and as such rely on clinical colleagues to report suspected TTI.” Additionally, failure to culture implicated platelet units, failure to perform blood cultures at the appropriate time and the transient nature of bacteremia result in underreporting of cases by passive surveillance. In an analysis of recent UK experience with large volume, delayed sampling (LVDS) primary culture, Benjamin et al. noted that the decline in bacterial contamination in the UK using LVDS primary culture from 2011 was similar to that found in the US with its lower volume, earlier testing protocol (67% versus 72.5%) following introduction of primary culture in the US in 2004 (41).
Hemovigilance data from the US National Healthcare Safety Network (NHSN) monitoring infections associated with 1,536,115 platelet transfusions, 2010–2016, in 308 facilities has recently been published (13). Thirty septic transfusion reactions associated with bacterial contamination were documented (19.5 per million platelet transfusions). Twenty-six resulted from 1,069,854 apheresis transfusions (24 per million), with one fatal reaction (coagulase-negative staphylococcus) and 3 life-threatening reactions (Acinetobacter baumannii, Escherichia coli and Staphylococcus aureus). Four resulted from 466,261 whole-blood derived platelet transfusions (8.6 per million), with one fatal reaction (Staphylococcus aureus) and two life-threatening reactions (Ralstonia pickettii and Staphylococcus aureus). The rate of septic reactions from apheresis platelets in this report (24 per million) is considerably higher that rates reported from passive surveillance programs (1–10 per million), but lower than those from active surveillance programs (36–50 per million), reflecting the limitations of passive surveillance programs and the mixed nature of these programs at institutions participating in NHSN.
These differences in septic transfusion rates based on active versus passive surveillance methods, as well as changes in presentation of transfusion reactions after introduction of primary culture (12), therefore considerably limit the value of assessing interventions based on septic transfusion reaction rates determined from passive surveillance data.
Incidence of bacterial contamination of platelet products
As the aim of interventions is to reduce the incidence of bacterial contamination, the most obvious and logical way to do this is to test platelet products at time of use by culture or direct bacterial detection method to document whether they are sterile or contaminated. Nine such studies have been performed on platelet units released as negative by primary culture (16,27,28,37,47-52). Methods used in these studies included BacT/ALERT culture bottles (n=6), plate culture (n=1), PDG Test (n=1) and PCR (n=1). Three of these studies were intervention studies using the PGD Test, PCR and culture, allowing contaminated units to be interdicted before use (27,28,49). All these studies showed remarkably similar contamination rates, ranging from 217 to 823 per million units, with overlapping 95% CIs, with the largest studies showing rates of 306–408 per million units. Contamination rates were not related to volume cultured; no information could be obtained on timing of primary culture as all cultures were performed within 24 h of collection.
Efficacy of primary culture strategies to reduce bacterial contamination
Prior to use of primary testing, the incidence of bacterial contamination at time of issue was approximately 400 per million apheresis units transfused (1:2,500) and 2,400 per million at-issue pooled units (1:417) (26). Associated septic transfusion rates (based on active bacterial surveillance) were 79 and 826 per million transfusions, respectively, with septic reactions occurring in approximately half of the patients receiving platelets with bacterial loads of >105 CFU/mL (approximately 50% of contaminated units had loads of >105 CFU/mL at time of transfusion). These “baseline” rates will be compared to rates reported with use of various interventions:
- Primary culture using 4–10 mL inoculated into one BacT/ALERT aerobic bottle or 3-4 mL inoculated into one eBDS culture pouch 24 h after collection. Using simple plate culture at time of issue, bacterial contamination (393 vs. 387/million; P=0.9) and active surveillance septic reaction rates (79 vs. 90/million; P=0.8) were unchanged for apheresis platelets, but both rates decreased significantly in WBD pools (2,415 vs. 198, P<0.0001 and 826 vs. 50/million, P<0.0001) (12). Contamination rates were comparable between platelets initially tested by suppliers using BacT/ALERT aerobic bottles and eBDS pouches.
- Primary culture of apheresis collections using two 4–5 mL aliquots inoculated into one BacT/ALERT aerobic and one anaerobic bottle 24–36 h after collection (PASSPORT study). This study has been reviewed earlier in this review and showed that use of 8–10 mL of platelet collection products with both aerobic and anaerobic culture bottles (in contrast to use of aerobic culture systems in I above) was not associated with a lower residual bacterial contamination rate (662 per million) (16), and the study was deemed a failure on this basis.
- Minimal proportional sampling volume (MPSV). Primary culture was studied on Trima leukoreduced apheresis platelets collected during two consecutive study periods using BacT/ALERT aerobic culture bottles inoculated from the mother bag 24 to 36 hours after collection (46). Period A: 8‐mL was inoculated into in one bottle; Period B: 3.8% of the mother bag, referred to as the “minimal proportional sample volume (MPSV)” method, 7–30 mL, was inoculated into one to three bottles (7–10 mL per bottle). True positive primary culture rates were: Period A, 90 per million out of 188,389 collections; Period B, 183 per million out of 159,098 collections (P<0.05). One septic transfusion reaction was reported in each period, which works out, using a 1.8 split ratio of collections into units, to septic reaction rates of 2.9 and 3.5 per million transfusions, respectively. The authors concluded that they had shown that the MPSV method improved the sensitivity of primary testing, and that it may represent another approach to enhanced safety for 5‐day storage without a requirement for secondary testing. The evidence presented certainly supported the first conclusion (improved sensitivity of primary testing using MPSV) but not the second conclusion that using MPSV enhanced safety (it did not—septic reactions were comparable and were very low, indicating again the limitations of passive surveillance). Furthermore, while no information on secondary culture testing was included in the publication, limited secondary testing of Period B units, was in fact performed and reported in abstract form on 8,039 outdated units, showing a contamination rate of 373–746 per million (the lower value using true positive results and the higher value including indeterminate results) (51,52). These contamination rates are no different from findings in many other studies where primary testing was performed using 4–8 mL volumes as well as the PASSPORT study. This study therefore provided no evidence of improved safety or that secondary testing is not needed, and this method, initially included in the draft guidance, was not included in the final FDA guidance document (14,15).
- Large volume, delayed sampling (LVDS). Primary culture of platelets was introduced in the UK by National Health Service Blood and Transplant (NHSBT) in 2011 (48). This program was instituted on buffy coat pools and split components of apheresis collections, in contrast to most other studies where apheresis collections are tested before being split into unit doses. Platelet components were sampled at 36 to 48 hours after donation and tested in BacT/ALERT aerobic and anaerobic bottles, with 8 mL inoculated into each bottle, leading to this study being designated LVDS, with approximately 7% of platelet volume cultured, and tested platelets were given a 7-day shelf-life. Results for 1,239,029 components tested to September 2015 were published (48), with the findings subsequently updated to March 2018, with a total of 1.3 million apheresis units and over 600,000 buffy coat pools tested (21). Study findings included a true positive detection rate from primary testing of 0.03% (300 per million). One septic transfusion reaction was reported due to S. aureus, with four “near misses” (contaminated units interdicted due to visible clots in units), three with S. aureus and one with Serratia marcescens. An additional 4 “near misses” have subsequently been detected (41,43). Secondary culture was performed on 6,217 outdated apheresis and buffy coat pool products, with one confirmed positive (161 per million) associated with Streptococcus pneumoniae in an apheresis unit. The authors concluded that the implementation of their bacterial screening protocol was an effective risk reduction measure and increased the safety of the blood supply. Critical analysis of this study, as with the MPSV study above, shows that the study lacked clearly defined goals and measurable endpoints. The primary culture positivity rate (300 per million) was comparable to the mean of 230 per million reported in a meta-analysis study (8) and 231 per million in the PASSPORT study (16). The major issue with assessment of this study is not whether it was beneficial (it was as no primary testing was in use prior to the study), but
was it more beneficial than standard primary culture? Comparison of at-issue or outdate culture results of each platelet type before and after institution of this program would readily and inexpensively provide the answer. The authors of the LVDS study have information on outdate culture results for each platelet type before institution of their primary culture program, and need to perform more cultures with use of their primary culture protocol as these findings will be very helpful in determining the changes associated with the two platelet types used. The information provided—1 positive out of 6,217 outdated apheresis and buffy coat pool products (161 per million)—is insufficient to determine if LVDS has any merit. Depending on the positivity rate of outdate or at-issue cultures, up to 50,000 units will need to be cultured to demonstrate superior performance of the LVDS method (12). Finding one positive among the 6,217 outdated units tested (a rate of 161 per million) is a failure rate comparable to that of the PASSPORT study.
A recently published study of a modified LVDS method performed 2017–2019 at Canadian Blood Services showed findings from primary culture performed ≥36 hours after collection (53). Mother bags of double apheresis collections were cultured by inoculation of 32 mL divided between three aerobic and one anaerobic bottles, while single unit apheresis and 4-unit buffy coat pools were cultured by inoculation of 16 mL divided into one aerobic and one anaerobic bottle; culture-negative units were released after incubation for 6 h with a 7-day outdate. Just over 75,000 apheresis units were transfused and outdate cultures, mostly tested on Day 8, were performed on 2,356 units, with two true positives, a rate of 849 (95% CI: 263–3,062) per million that was no different from the apheresis unit outdate culture rate at the same center with low volume culture (8–10 mL per apheresis collection at ≥24 hours after collection) of 941 (95% CI: 484–1,854) per million (8/8,498) (47). Contamination rates of outdated buffy coat pools were 7/8,535 (820 per million, 95% CI: 405–1,689) pre-LVDS and 4/2,954 (1,345 per million, 95% CI: 550–3,462) with LVDS, again showing overlapping confidence intervals. As with the LVDS testing performed by NHSBT discussed above, these high rates are of concern and much larger sample sizes are needed.
These issues associated with LVDS were discussed at the 2017 and 2018 BPAC meetings, during both of which one of the committee members commented that the data presented did not support use of this strategy as there are limited gains from the increases in volume and later timing of cultures (21,54). Similar inconclusive findings about the value of LVDS performed 48 h after collection in Quebec, Canada were presented in 2018, where 5/9,165 contaminated units cultured at outdate (545 per million) were found prior to introduction of LVDS and 0/3,185 with LVDS, an insufficient sample size to show statistical significance (55).
Efficacy of secondary testing
Secondary testing is generally performed on or after day 3. A variety of secondary testing methods have been described, including “rapid” tests (27,28,29,56,57) which have a lower limit of detection of 103 to 105 CFU/mL and secondary culture (49,50) which has a lower limit of detection of <102 CFU/mL.
Rapid testing
While several methods have been developed, the only rapid testing method currently marketed is the PGDprime Test (Verax Biomedical), previously available as the PGD Test. This test is a simple, rapid, day of transfusion test for the detection of bacterial contamination in platelets and is based on Pan Genera Detection (PGD) technology (17). It detects the presence of conserved antigens lipoteichoic acid and lipopolysaccharide found on aerobic and anaerobic Gram positive and Gram negative bacteria, respectively. A multicenter study of the PGD Test documented detection of nine bacterially contaminated apheresis units out of 27,620 units tested, a rate 326 per million (27). A more recent report on the experience of six institutions performing rapid testing of apheresis units tested on day 6 or day 7 to extend outdate documented detection of 9 true positive rapid tests out of 6,556 units tested, a rate of 1,374 per million (95% CI: 732–2,604) (58). This rate had overlapping 95% CIs with outdate cultures (7-day outdate in this study) in the PASSPORT study (4/6,039, 662 per million, 95% CI: 269–1,695) (16) and >6 day outdate cultures in the product label of BacT/ALERT bottles (46/32,142, 1,431 per million, 95% CI: 1,075–1,908) (18). These high rates in older units are a major concern, and rapid testing has the advantage of preventing, rather than just documenting, bacterial contamination. Over 1.4 million PGD tests have been performed, with no reports of fatalities and, for the period 2015–2019, two reports of septic transfusion reactions associated with the same apheresis collection due to Acinetobacter baumannii-calcoaceticus, which was not detected by the PGD Test as it does not detect this species (59). The reader is referred to a companion article in this series on this product for further information (60).
Secondary culture
Culture later in the shelf life of platelets has been shown to be very effective due to the time delay allowing sufficient time for small inocula to grow, combined with the very low inoculum needed to show detection by culture. A study of 43,230 platelet units performed by the Irish Blood Transfusion Service in 2005 showed that primary aerobic and anaerobic culture detected 27% of contaminated units, while secondary culture on day 4 detected 41% and outdate culture the remaining 32% of contaminated units (50). This study and subsequent testing by the Irish Blood Transfusion Service was presented at two of the recent BPAC meetings and appears to be the basis of the recommendation for the day 4 secondary culture in the guidance (21,54).
Another secondary culture study performed on day 3 at Johns Hopkins Hospital and Medical Institutions has been published and was also presented at these BPAC meetings (49). In this study, performed 2016–2017, 23,044 platelet products were sampled using 5 mL inoculated into aerobic bottles. Eight positive cultures were detected and interdicted, seven of which were positive within 24 hours, with five of the eight cases positive on repeat culture, an incidence of 217 per million. No septic transfusion reactions were reported during the observation period, and the cost per averted case was $77,935. This study appears to be the basis of the recommendation for the day 3 secondary culture in the guidance (21,54). This study has now been updated to show results on 55,896 platelet units tested 2016–2019, 81% using 5 mL and 19% using 10 mL culture aliquots, with no septic reactions during the study period (61). Thirty secondary cultures were positive, with 14 true positives (250 per million)—12/45,251 in the 5 mL group (265 per million) and 2/10,645 in the 10 mL group (188 per million, P=0.65). The rate of false positives was significantly higher in the 10 mL group (7 vs. 1, P<0.0001).
The FDA guidance states that “Bacterial testing to extend dating beyond day 5 and up to day 7 should be performed with devices labeled with LVDS as an acceptable safety measure.” FDA has cleared BacT/ALERT aerobic and anaerobic bottles with this “safety measure” claim, which is defined as testing that can be used to extend dating of platelets provided that sampling should be done no sooner than day 4 post collection with both aerobic and anaerobic bottles and with 8–10 mL sample per bottle, and that negative results from days 4–6 of safety measure testing can be used to extend the dating of platelets to 7 days using approved storage bags (18). The basis for this safety claim is shown in the product labeling under “Performance of the BacT/ALERT 3D Systems for Use as a Secondary Safety Measure Test to Extend the Shelf Life of Platelet Preparations.” Data are presented based on a literature review and data from blood collection and transfusion services that used the BacT/ALERT 3D system for secondary testing and/or end-date QC surveillance of previously tested platelets. Data were available on a total of 128,124 leukocyte-reduced apheresis units that were determined negative during quality control testing and released for transfusion that were tested on days 3, 4, and ≥6 days post collection. A total of 72 positive bottles (0.06%) were detected (data sources are not provided)—a rate of 562 (95% CI: 447–708) per million. More relevant to secondary testing performed to extend the shelf-life of platelets is the analysis provided on day 3 and day 4 units. The rate was 309 (95% CI: 145–673) per million on day 3 units and 261 (95% CI: 170–403) per million on day 4 units, showing comparable detection to rapid testing (326 per million, 95% CI: 149–618 per million), the first method to obtain “safety measure” labeling (17,27). As discussed under rapid testing above, the considerably higher contamination rate of ≥6 days units by secondary culture of 1,431 per million units is a major concern about the safety of transfusion of these older units without secondary testing.
Implementation of secondary culture by hospital Transfusion Services is a complex undertaking, requiring acquisition of or access to instruments for incubation of bottles (BacT/ALERT 3D, BacT/ALERT Virtuo or other blood culture systems), a laminar flood hood for sampling platelet units, sterile connection devices and sampling kits, and syringes and needles to inoculate culture bottles, as well as facilities to investigate positive bottles (48-50). Use of a device labeled as a “safety measure” is not required when secondary testing is performed to adequately control bacterial risk through day 5 of storage.
Efficacy of pathogen reduction
The Intercept system for pathogen reduction (Cerus Corporation) has been used to treat over 1.1 million platelet units in Europe and over 500,000 in the US, with a projected use in the US of >400,000 apheresis units in 2021. Two transfused apheresis units contaminated with Acinetobacter calcoaceticus-baumannii have been reported in the US and resulted in severe septic reactions, with a fatal outcome in one patient (62,63). These two contaminated units were separated in time and location, with no obvious common source despite the isolates being clonal with each other as well as with two other geographically distinct contaminated, but not pathogen reduced, apheresis units (25,59,64,65). Based on the clonal nature of the isolates and the fact that the pathogen reduction process has been shown to be effective against these isolates, it appears likely that contamination of units occurred after the pathogen reduction process had been performed and that a common source is involved. In the most recent case a defect in the apheresis bag was detected, suggesting that the contaminating bacteria were in fact introduced after the pathogen reduction process had been performed (25). The reader is referred to a recent review of pathogen reduction for further information on this technology (66).
FDA guidance
This guidance, titled “Bacterial Risk Control Strategies for Blood Collection Establishments and Transfusion Services to Enhance the Safety and Availability of Platelets for Transfusion; Guidance for Industry” was finally issued in September 2019 after being in draft form since 2014 and the subject of three Blood Product Advisory Committee meetings (15,21,54). Recommended implementation date is no later than 18 months after the guidance issue date, which is March 2021; this implementation date has subsequently been extended by 6 months due to the SARS-Covid-19 pandemic.
The guidance consists of a cover page, a table of contents (page i) and contents (pages 1–19). All pages except the cover page include the header “Contains Nonbinding Recommendations”, and page 1 starts with the following text in a black box:
“This guidance represents the current thinking of the Food and Drug Administration (FDA or Agency) on this topic. It does not establish any rights for any person and is not binding on FDA or the public. You can use an alternative approach if it satisfies the requirements of the applicable statutes and regulations. To discuss an alternative approach, contact the FDA staff responsible for this guidance as listed on the title page.”
This is followed by an introduction, which includes the following disclaimer:
“FDA’s guidance documents, including this guidance, do not establish legally enforceable responsibilities. Instead, guidances describe the FDA’s current thinking on a topic and should be viewed only as recommendations, unless specific regulatory or statutory requirements are cited. The use of the word should in FDA’s guidances means that something is suggested or recommended, but not required.”
Background information follows, stating that the risk of bacterial contamination of platelets is a leading risk of infection from blood transfusion, estimated at about 1:2,500 transfusions, and that fatal transfusion reactions from undetected contaminated platelet collections continue to occur despite the implementation of numerous interventions, including the commonly used method of a single culture performed no sooner than 24 hours after collection of the platelets. Reported rates of septic transfusion reactions vary from 1/100,000 by passive surveillance to 1/10,000 by active surveillance. FDA has established regulations to address the control of bacterial contamination of platelets such as 21 CFR 606.145(a), which state that blood establishments and transfusion services must assure that the risk of bacterial contamination of platelets is adequately controlled using FDA approved or cleared devices, or other adequate and appropriate methods found acceptable for this purpose by FDA. Since 2012, FDA has held three Blood Products Advisory Committee (BPAC) meetings to address bacterial risk in platelet products. The most recent BPAC meeting, in July 2018, discussed the scientific evidence and operational considerations of available strategies to control the risk of bacterial contamination of platelets with 5- and 7-day dating, including bacterial testing strategies (using culture-based and rapid bacterial detection devices) and pathogen reduction technology. Subsequently, FDA published the December 2018 draft guidance. Comments to the published draft guidance documents, as well as the BPAC 2018 proceedings, provided the foundation for the recommendations in this guidance, published in September 2019 and updated in December 2020.
The recommendations section of the guidance start on page 3, and are summarized below.
General considerations
- The recommendations in this guidance entail the use of FDA-cleared or approved bacterial detection devices, pathogen reduction devices, and platelet storage containers.
- Bacterial detection testing, pathogen reduction, and storage in platelet containers must be conducted consistent with the instructions for use of the device [21 CFR 606.65(e)].
- Blood collection establishments and transfusion services should have in place measures to promptly alert the collection establishment or transfusion service if a distributed platelet product is subsequently identified as positive for bacterial contamination.
- Depending on the recommendation, sampling or testing time in this guidance is expressed in units of hours or days:
- When timing is expressed in units of hours, it is in reference to the actual hour of collection or sampling. For example, if a platelet product is collected at 9 am (0:900) on January 1st, sampling performed no earlier than 24 hours from time of collection means that sampling should be performed no earlier than 9 am (0:900) on January 2nd; sampling performed no earlier than 36 hours means sampling no earlier than 9 pm (21:00) on January 2nd.
- When timing is expressed in units of days, it is in reference to the day of collection, which is considered day 0. A day is defined as beginning at midnight (00:00) and ending at 23:59. Expiry day refers to 23:59 of the stated day (i.e., prior to midnight). For example, if a platelet is collected on January 1st (day 0) at 9 am (0:900) with a 5-day storage period, it means it will expire at 23:59 on January 6th.
- When sampling is recommended to be conducted on a specific day, it means that sampling can occur at any time on that day prior to midnight, regardless of the time of the initial collection. For example, if a platelet is collected on January 1st at 9 am (day 0), secondary culture performed on day 3 should be performed on January 4th, at any time prior to midnight.
- Products may be shipped during the recommended culture incubation periods, provided the blood collection establishment establishes procedures to maintain control of the product during the incubation period.
- Blood collection establishments and transfusion services must not release for transfusion platelets identified as bacterially contaminated [21 CFR 606.145(b) and (c)].
- Following secondary testing, it is not expected that blood collection establishments or transfusion services retest units to determine platelet yield.
- Blood collection establishments and transfusion services should establish procedures to assure traceability of the bacterial testing status of platelet products in their inventory.
Strategies for testing of apheresis and WBD pre-storage pools
Single-step strategies
There are four single-step options, three requiring culture and one pathogen reduction. The culture options all specify minimum culture volumes of 8 mL in an aerobic bottle and 8 mL in an anaerobic bottle per apheresis unit or WBD pool, with a minimum incubation period of 12 h before release. The three culture options are shown in Table 2 and include testing units ≥36 h after collection (5-day shelf-life), ≥48 h after collection (7 day shelf-life), and ≥24 h after collection (3-day shelf-life); the last option also allows culture of apheresis collections (mother bags). The fourth option is pathogen reduction of apheresis units ≤24 h after collection, with a shelf-life of 5 days.
Table 2
| Strategy | Primary culture minimum volume per bottle (bottle type) | Time after collection (hours) | Incubation time before release (hours) | Shelf life (days) | Shelf life extension allowed with second step |
|---|---|---|---|---|---|
| Culture at ≥36 h after collection of split apheresis units or pre-storage derived pools | 8 mL (aerobic) and 8 mL (anaerobic) per unit or pool | ≥36 | 12 | 5 | Apheresis units only to 7 days |
| Culture at ≥48 h after collection of split apheresis units | 8 mL (aerobic) and 8 mL (anaerobic) per unit | ≥48 | 12 | 7 | NA |
| Culture at ≥24 h after collection of apheresis collections (mother bags), apheresis split units or pre-storage pools | 8 mL (aerobic) and 8 mL (anaerobic) per collection, unit or pool | ≥24 | 12 | 3 | Pools and apheresis in PAS to 5 days. Apheresis in plasma to 7 days |
| Pathogen reduction of apheresis units within 24 h of collection | NA | ≤24 | NA | 5 | No |
*, information from (15). Guidance specifies that times in hours are defined as exact time after collection or sampling, while times in days are defined as any time on day specified.
Two-step strategies
There are four pathways available, depending on which single-step strategy is used, with secondary testing being either culture or rapid test. Rapid test refers to the PGDprime Test (Verax Biomedical, Marlborough, MA), which is the only rapid test currently available and which has the required “Safety Measure” label cleared by FDA. These pathways are shown in Figure 1 and include primary culture performed ≥24 h after collection and secondary culture of 8 mL in an aerobic bottle performed on day ≥3 (extends shelf-life from 3 to 5 days); primary culture performed ≥36 h after collection and secondary culture performed on day ≥4 (extends shelf-life from 5 to 7 days); primary culture performed ≥24 h after collection and secondary testing by rapid test within 24 h of transfusion on day ≥3 (extends shelf-life from 3 to 5 or 7 days; and primary culture performed ≥36 h after collection and secondary testing by rapid test within 24 h of transfusion on day ≥3 (extends shelf-life from 5 to 7 days).
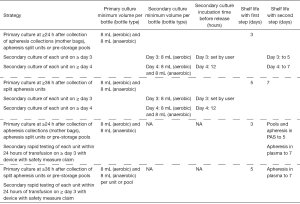
Strategies for testing of single WBD units and post-storage WBD pools
These platelet types have a 5-day shelf-life, and two testing strategies are available—rapid testing and culture (Table 3). Rapid testing can be performed on single WBD units and post-storage WBD pools within 24 h of transfusion, with post-storage pools expiring 4 hours after pooling. Culture of single WBD units can be performed ≥24 h after collection in an aerobic bottle, using the largest practical volume within the range permitted by the bacterial testing device used due to the small volume of these units.
Table 3
| Strategy | Primary culture minimum volume per bottle (bottle type) | Incubation time before release (hours) | Shelf life (days) |
|---|---|---|---|
| Rapid testing of single WBD units within 24 hours of transfusion with device with safety measure claim | NA | NA | 5 |
| Rapid testing of post-storage WBD pools at time of pooling with device with safety measure claim+ | NA | NA | 5 |
| Culture of single WBD units ≥24 h after collection | Largest practical volume (aerobic) | 12 | 3# |
| Culture of single WBD units ≥36 h after collection | Largest practical volume (aerobic) | 12 | 5 |
*, information from (15). Guidance specifies that times in hours are defined as exact time after collection or sampling, while times in days are defined as any time on day specified; +, post-storage WBD pools expire 4 hours after pooling; #, can be extended to 5 days with rapid testing.
Additional requirements
The guidance document includes a number of administrative items that are required with implementation of various strategies, including requirements for labeling and dating platelet products based on primary and secondary testing, obtaining appropriate manufacturing licensing for interstate commerce, validation plans and quality control, annual reports, registration of transfusion services as engaged in the manufacture of blood products if bacterial detection devices are used to extend outdate to 6 or 7 days, and finally that the recommended period of implementation of the guidance is no later than 18 months after the guidance issue date.
Platelet availability implications of FDA guidance strategies
Platelets are a scarce resource and currently ~300,000 units outdate annually in the US. All primary culture strategies in the new guidance have very specific timing requirements for performance after collection that will result in platelets being released up to a day later than under current practice, where primary cultures are frequently performed <24 h after collection, with substantial variation in the minimum incubation time of cultures before release (67). Introduction of LVDS with extension of outdate from 5 to 7 days by one Canadian supplier only reduced outdating by 31% (53). Use of the LVDS strategy with a 5 day outdate will increase rather that reduce the outdate rate without the use of secondary testing.
Financial implications of FDA guidance strategies
The mean cost of apheresis units in 2017 was $522 per unit (9). Major blood product suppliers have advised hospitals that the incremental cost of LVDS units will be around $80 per unit, while that of pathogen reduced units will be $150 per unit, bringing totals to $602 and $672, respectively. A recent analysis of the financial impact of the various strategies projected incremental mean costs per unit of $31.93 for secondary rapid testing, $17.26 for 5-day shelf-life secondary culture and $44.60 for 7-day shelf-life secondary culture (68). Incremental cost of LVDS units was estimated at $30 per unit (now known to be $80 per unit), and further calculations showed that LVDS would need to cost $22.32 to be cheaper per transfusion than all other strategies and less than $32.02 to be cheaper than 7-day shelf-life secondary culture. Based on platelet supply projections for the near future being evenly divided into pathogen reduced and 5-day outdate LVDS tested units, with one million of each type used per year, the annual incremental costs would be $150 million for pathogen reduced and $80 million for LVDS units. In contrast, the incremental cost of secondary testing would be $17.26 million for secondary culture and $31.93 million for secondary rapid testing per million platelet units. Furthermore, secondary testing would extend shelf-life to 7 days, which would more than mitigate the cost of secondary testing. A recent survey of 66 hospitals using rapid testing documented overall savings on platelet acquisition costs due to virtual elimination of wastage of $80 per unit amortized over all platelet units, which more than compensated for the modest cost of rapid testing of $5.29 per unit amortized over all platelet units (amortization based on secondary testing on 16.5% of platelet units costing $485 per apheresis unit and rapid test costing $32 per unit tested) (60,69). While the high cost of pathogen reduced units may be justified by safety considerations, this is not the case with LVDS units, for which the incremental safety improvement is not known as discussed earlier.
Advantages and limitations of FDA Guidance
As discussed in the introduction, there is evidence supporting the efficacy of secondary testing by culture or rapid testing, and use of these methods should be encouraged. Secondary culture offers the advantage of broader spectrum of organism coverage and lower detection limit, while rapid testing offers the obvious advantage of speed, with turn-around time of a batch of rapid tests taking 30 minutes to perform. The disadvantage of secondary testing is that products need to be tested by the end user or returned to the supplier for testing; both options present cost and logistic issues.
As also extensively discussed, at this time there is insufficient evidence showing that LVDS is superior to current primary culture protocols, with further data from outdate or at-issue cultures needed. It needs to be pointed out that the efficacy of current primary culture protocols was not known at this time this was introduced, and we now know that they need improvement, after they have been in use for 15 years. While LVDS has the potential for avoiding the need for secondary testing, we need to know this rather than assume that this is the case. It is also of particular concern that, in view of the lack of adequate evidence of efficacy, the FDA guidance does not include a requirement to perform the studies needed to show efficacy, as was required, for example, for culture bottles used for primary and secondary culture methods before they could be implemented (18). It is particularly concerning as the incremental cost of performing LVDS on one million apheresis units will be $80 million.
Conclusions
Among the strategies included in the FDA guidance document, four single-step and four two-step pathways are provided for apheresis and pre-storage WBD pools. Three of the single-step pathways are based on primary culture of 16 mL performed 24 h after collection on mother bags, split apheresis units and WBD pools or 36–48 h after collection on split apheresis units. The fourth single-step pathway is pathogen reduction performed within 24 h of collection. Shelf-life for these single-step pathways range from 3–7 days. All of the two-step pathways require primary culture of at least 16 mL per collection at 24 h or per unit 24–36 h after collection, with secondary testing by culture or rapid test; secondary testing extends shelf-life to up to 7 days. Two pathways are provided for single and post-storage pooled WBD units—rapid testing for both platelet types and culture at 24–36 h for single units. There is evidence that all these pathways reduce the incidence of bacterial contamination of platelet products, except for the 36 h and 48 h single-step culture pathways, where additional evidence of efficacy is needed. The cost of these new strategies is high while the proven efficacy of these strategies varies considerably.
Acknowledgments
The contribution of the late Roslyn A. Yomtovian, MD, Medical Director, Transfusion Service, University Hospitals Cleveland Medical Center and Case Western Reserve University School of Medicine, Cleveland, Ohio, 1988–2007, to the field of bacterial contamination of platelets is recognized with appreciation.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Sandra Ramirez-Arcos) for the series “Bacterial Contamination of Platelet Components” published in Annals of Blood. The article was sent for external peer review organized by the Guest Editor and the editorial office.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aob-21-10). The series “Bacterial Contamination of Platelet Components” was commissioned by the editorial office without any funding or sponsorship. Dr. MRJ reports grants and personal fees from bioMérieux, grants from Charles River Labs, grants from Fenwal, grants from Gambro, grants from Genprime, grants from Hemosystem, grants and personal fees from Immunetics, grants from Pall, grants and personal fees from Verax, other from Biosense, other from Lyntech, other from Blood Systems, during the conduct of the study. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jacobs MR, Good CE, Lazarus HM, et al. Relationship between bacterial load, species virulence, and transfusion reaction with transfusion of bacterially contaminated platelets. Clin Infect Dis 2008;46:1214-20. [Crossref] [PubMed]
- Buchholz DH, Young VM, Friedman NR, et al. Bacterial proliferation in platelet products stored at room temperature. Transfusion-induced Enterobacter sepsis. N Engl J Med 1971;285:429-33. [Crossref] [PubMed]
- Brecher ME, Blajchman MA, Yomtovian R, et al. Addressing the risk of bacterial contamination of platelets within the United States: a history to help illuminate the future. Transfusion 2013;53:221-31. [Crossref] [PubMed]
- Eder AF, Kennedy JM, Dy BA, et al. Bacterial screening of apheresis platelets and the residual risk of septic transfusion reactions: the American Red Cross experience (2004-2006). Transfusion 2007;47:1134-42. [Crossref] [PubMed]
- Benjamin RJ, Kline L, Dy BA, et al. Bacterial contamination of whole-blood-derived platelets: the introduction of sample diversion and prestorage pooling with culture testing in the American Red Cross. Transfusion 2008;48:2348-55. [Crossref] [PubMed]
- Holme S, McAlister MB, Ortolano GA, et al. Enhancement of a culture-based bacterial detection system (eBDS) for platelet products based on measurement of oxygen consumption. Transfusion 2005;45:984-93. [Crossref] [PubMed]
- Eder AF, Kennedy JM, Dy BA, et al. Limiting and detecting bacterial contamination of apheresis platelets: inlet-line diversion and increased culture volume improve component safety. Transfusion 2009;49:1554-63. [Crossref] [PubMed]
- White SK, Schmidt RL, Walker BS, et al. Bacterial contamination rate of platelet components by primary culture: a systematic review and meta-analysis. Transfusion 2020;60:986-96. [Crossref] [PubMed]
- Jones JM, Sapiano MRP, Savinkina AA, et al. Slowing decline in blood collection and transfusion in the United States - 2017. Transfusion 2020;60:S1-S9. [Crossref] [PubMed]
- Savinkina AA, Haass KA, Sapiano MRP, et al. Transfusion-associated adverse events and implementation of blood safety measures - findings from the 2017 National Blood Collection and Utilization Survey. Transfusion 2020;60:S10-S16. [Crossref] [PubMed]
- Sapiano MRP, Jones JM, Savinkina AA, et al. Supplemental findings of the 2017 National Blood Collection and Utilization Survey. Transfusion 2020;60:S17-S37. [Crossref] [PubMed]
- Kundrapu S, Srivastava S, Good CE, et al. Bacterial contamination and septic transfusion reaction rates associated with platelet components before and after introduction of primary culture: experience at a US Academic Medical Center 1991 through 2017. Transfusion 2020;60:974-85. [Crossref] [PubMed]
- Haass K, Sapiano M, Savinkina A, et al. Transfusion-transmitted infections reported to the National Healthcare Safety Network Hemovigilance Module. . Transfusion Medicine Reviews 2019. Available online: https://www.sciencedirect.com/science/article/pii/S0887796318301044
- CBER-FDA. Bacterial Detection Testing by Blood Collection Establishments and Transfusion Services to Enhance the Safety and Availability of Platelets for Transfusion. Available online: https://www.regulations.gov/document/FDA-2014-D-1814-0001 2014:Docket Number: FDA-2014-D-1814.
- CBER-FDA. Bacterial Detection Testing by Blood Collection Establishments and Transfusion Services to Enhance the Safety and Availability of Platelets for Transfusion. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/bacterial-risk-control-strategies-blood-collection-establishments-and-transfusion-services-enhance 2020:Docket Number: FDA-2014-D-1814.
- Dumont LJ, Kleinman S, Murphy JR, et al. Screening of single-donor apheresis platelets for bacterial contamination: the PASSPORT study results. Transfusion 2010;50:589-99. [Crossref] [PubMed]
- CBER-FDA. Platelet PGD Test System: BK160087. Available online: https://www.fda.gov/vaccines-blood-biologics/substantially-equivalent-510k-device-information/platelet-pgd-test-system-bk160087 2017.
- CBER-FDA. BK170142: BacT/ALERT BPA Culture Bottle; BacT/ALERT BPN Culture Bottle. Available online: https://www.fda.gov/vaccines-blood-biologics/substantially-equivalent-510k-device-information/bk170142-bactalert-bpa-culture-bottle-bactalert-bpn-culture-bottle 2018.
- CBER-FDA. BK180211: BD BACTEC Platelet Aerobic/F Culture vials; BD BACTEC Platelet Anaerobic/F Culture Vials. Available online: https://www.fda.gov/vaccines-blood-biologics/substantially-equivalent-510k-device-information/bk180211-bd-bactec-platelet-aerobicf-culture-vials-bd-bactec-platelet-anaerobicf-culture-vials 2018.
- Chetouane Y, Gallian P, Chetouane K, et al. Comparing two blood culture systems for the detection of bacterial contamination in platelet concentrates. Transfusion 2018;58:2604-10. [Crossref] [PubMed]
- CBER-FDA. Transcript of FDA CBER Blood Products Advisory Committee Meeting, FDA White Oak Campus, Silver Spring, MD; July 18, 2018. Available online: https://www.fda.gov/media/115633/download 2018.
- FDA Center for Biologics Evaluation and Research. Fatalities Reported to FDA Following Blood Collection and Transfusion: Annual Summary for Fiscal Year 2016. Available online: https://www.fda.gov/media/111226/download 2017.
- Horth RZ, Jones JM, Kim JJ, et al. Fatal Sepsis Associated with Bacterial Contamination of Platelets - Utah and California, August 2017. MMWR Morb Mortal Wkly Rep 2018;67:718-22. [Crossref] [PubMed]
- Jones J. Call for Cases: Acinetobacter Calcoaceticus-Baumannii Complex Sepsis Associated with Platelet Transfusion — Multi-state, 2018. Available online: https://www.cdc.gov/mmwr/volumes/68/wr/mm6823a2.htm 2018.
- Fadeyi E, Wagner S, Goldberg C, et al. Fatal Sepsis Associated with a Storage Container Leak Permitting Platelet Contamination with Environmental Bacteria after Pathogen Reduction. Transfusion 2021;61:641-8. [Crossref] [PubMed]
- Yomtovian R, Lazarus HM, Goodnough LT, et al. A prospective microbiologic surveillance program to detect and prevent the transfusion of bacterially contaminated platelets. Transfusion 1993;33:902-9. [Crossref] [PubMed]
- Jacobs MR, Smith D, Heaton WA, et al. Detection of bacterial contamination in prestorage culture-negative apheresis platelets on day of issue with the Pan Genera Detection test. Transfusion 2011;51:2573-82. [Crossref] [PubMed]
- Ladenheim H, Hibbard AJ, Weiss JW, et al. Detection of bacterial contamination in platelet components using an in-house developed real-time polymerase chain reaction amplification assay. Transfusion 2012;52:201A.
- Heaton WA, Good CE, Galloway-Haskins R, et al. Evaluation of a rapid colorimetric assay for detection of bacterial contamination in apheresis and pooled random-donor platelet units. Transfusion 2014;54:1634-41. [Crossref] [PubMed]
- Zaza S, Tokars JI, Yomtovian R, et al. Bacterial contamination of platelets at a university hospital: increased identification due to intensified surveillance. Infect Control Hosp Epidemiol 1994;15:82-7. [Crossref] [PubMed]
- Palavecino E, Yomtovian R. Risk and prevention of transfusion-related sepsis. Curr Opin Hematol 2003;10:434-9. [Crossref] [PubMed]
- Yomtovian R. Bacterial contamination of blood: lessons from the past and road map for the future. Transfusion 2004;44:450-60. [Crossref] [PubMed]
- Yomtovian R, Brecher ME. pH and glucose testing of single-donor apheresis platelets should be discontinued in favor of a more sensitive detection method. Transfusion 2005;45:646-8. [Crossref] [PubMed]
- Yomtovian RA, Palavecino EL, Dysktra AH, et al. Evolution of surveillance methods for detection of bacterial contamination of platelets in a university hospital, 1991 through 2004. Transfusion 2006;46:719-30. [Crossref] [PubMed]
- Palavecino EL, Yomtovian RA, Jacobs MR. Bacterial contamination of platelets. Transfus Apher Sci 2010;42:71-82. [Crossref] [PubMed]
- Hong H, Xiao W, Lazarus HM, et al. Detection of septic transfusion reactions to platelet transfusions by active and passive surveillance. Blood 2016;127:496-502. [Crossref] [PubMed]
- Kundrapu S, Jacobs MR, Maitta RW. Bacterial Contamination and Septic Transfusion Reaction Rates Associated with Platelet Components before and after Introduction of Primary Culture. Transfusion 2018;58 S2:217A.
- Srivastava S, Jacobs MR, Maitta RW. Comparative Efficacy of BacT/ALERT and eBDS Methods for Primary Culture of Platelet Products for Prevention of Bacterial Contamination of Platelet Components. Transfusion 2018;58 S2:204A.
- Srivastava S, Jacobs MR, Maitta RW. Differences in Bacterial Species and Loads between Platelet Components Tested by BacT/ALERT and eBDS Methods. Transfusion 2018;58 S2:216A.
- FDA. Bacterial Detection Testing by Blood Collection Establishments and Transfusion Services to Enhance the Safety and Availability of Platelets for Transfusion. Available online: https://wwwfdagov/media/123448/download 2020.
- Benjamin RJ, Braschler T, Weingand T, et al. Hemovigilance monitoring of platelet septic reactions with effective bacterial protection systems. Transfusion 2017;57:2946-57. [Crossref] [PubMed]
- Bolton-Maggs PH, Cohen H. Serious Hazards of Transfusion (SHOT) haemovigilance and progress is improving transfusion safety. Br J Haematol 2013;163:303-14. [Crossref] [PubMed]
- Narayan SE, Poles D, et al. on behalf of the Serious Hazards of Transfusion (SHOT) Steering Group. The 2019 Annual SHOT Report, 2020.
- Fuller AK, Uglik KM, Savage WJ, et al. Bacterial culture reduces but does not eliminate the risk of septic transfusion reactions to single-donor platelets. Transfusion 2009;49:2588-93. [Crossref] [PubMed]
- Erony SM, Marshall CE, Gehrie EA, et al. The epidemiology of bacterial culture-positive and septic transfusion reactions at a large tertiary academic center: 2009 to 2016. Transfusion 2018;58:1933-9. [Crossref] [PubMed]
- Kamel H, Townsend M, Bravo M, et al. Improved yield of minimal proportional sample volume platelet bacterial culture. Transfusion 2017;57:2413-9. [Crossref] [PubMed]
- Ramirez-Arcos S, DiFranco C, McIntyre T, et al. Residual risk of bacterial contamination of platelets: six years of experience with sterility testing. Transfusion 2017;57:2174-81. [Crossref] [PubMed]
- McDonald C, Allen J, Brailsford S, et al. Bacterial screening of platelet components by National Health Service Blood and Transplant, an effective risk reduction measure. Transfusion 2017;57:1122-31. [Crossref] [PubMed]
- Bloch EM, Marshall CE, Boyd JS, et al. Implementation of secondary bacterial culture testing of platelets to mitigate residual risk of septic transfusion reactions. Transfusion 2018;58:1647-53. [Crossref] [PubMed]
- Murphy WG, Foley M, Doherty C, et al. Screening platelet concentrates for bacterial contamination: low numbers of bacteria and slow growth in contaminated units mandate an alternative approach to product safety. Vox Sang 2008;95:13-9. [Crossref] [PubMed]
- Townsend J, Bravo M, Vanderpool M, et al. Surveillance cultures on day-7 apheresis platelets which outdated on day 5. Vox Sanguinis 2015;109:232.
- Vassallo RR. Presentation at FDA July 18-19, 2018 Blood Products Advisory Committee Meeting. Available online: https://www.fda.gov/media/115633/download 2018.
- Ramirez-Arcos S, Evans S, McIntyre T, et al. Extension of platelet shelf life with an improved bacterial testing algorithm. Transfusion 2020;60:2918-28. [Crossref] [PubMed]
- CBER-FDA. Transcript of FDA CBER Blood Products Advisory Committee Meeting, FDA White Oak Campus, Silver Spring, MD; November 30, 2017. Available online: https://www.fda.gov/media/109985/download 2017.
- Delage G. Delayed sampling, high volume bacterial culture of platelets: one center’s experience. Annual Meeting AABB; 2018; Boston.
- Jacobs MR, Bajaksouzian S, Windau A, et al. Evaluation of the Scansystem method for detection of bacterially contaminated platelets. Transfusion 2005;45:265-9. [Crossref] [PubMed]
- Alrabeh R, Korte L, Reyes M, et al. Long-term Experience with Rapid Screening for Platelet Bacterial Contamination in a High-volume Transfusion Service. Ann Clin Lab Sci 2019;49:748-55. [PubMed]
- Mintz PD, Sanders JR, Blair J, et al. Confirmed Positive Bacterial Detection in Platelet Concentrates by a Rapid Test after Negative Primary Culture. Transfusion 2019;59:66A.
- Jones SA, Jones JM, Leung V, et al. Sepsis Attributed to Bacterial Contamination of Platelets Associated with a Potential Common Source - Multiple States, 2018. MMWR Morb Mortal Wkly Rep 2019;68:519-23. [Crossref] [PubMed]
- Mintz PD. The PGD Verax Immunoassay for detection of bacterial contamination in platelet concentrates: Sensitivity and Specificity. Ann Blood 2021; [Crossref]
- Fenwick AJ, Gehrie EA, Marshall CE, et al. Secondary bacterial culture of platelets to mitigate transfusion-associated sepsis: A 3-year analysis at a large academic institution. Transfusion 2020;60:2021-8. [PubMed]
- MAUDE Adverse Event Report. CERUS CORPORATION INTERCEPT BLOOD SYSTEM FOR PLATELETS Severe septic transfusion reaction Acinetobacter baumanii. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfmaude/detail.cfm?mdrfoi__id=10292518&pc=PJF 2018.
- MAUDE Adverse Event Report. CERUS CORPORATION INTERCEPT BLOOD SYSTEM FOR PLATELETS Severe septic transfusion reaction due to Acinetobacter baumanii, Leclercia adecarboxylata, Staphylococcus saprophyticus. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfmaude/detail.cfm?mdrfoi__id=10292518&pc=PJF 2020.
- Fridey JL, Stramer SL, Nambiar A, et al. Sepsis from an apheresis platelet contaminated with Acinetobacter calcoaceticus/baumannii complex bacteria and Staphylococcus saprophyticus after pathogen reduction. Transfusion 2020;60:1960-9. [PubMed]
- Nevala-Plagemann C, Powers P, Mir-Kasimov M, et al. A Fatal Case of Septic Shock Secondary to Acinetobacter Bacteremia Acquired from a Platelet Transfusion. Case Rep Med 2019;2019:3136493. [Crossref] [PubMed]
- Seltsam A. Pathogen Inactivation of Cellular Blood Products-An Additional Safety Layer in Transfusion Medicine. Front Med (Lausanne) 2017;4:219. [Crossref] [PubMed]
- Brecher ME, Jacobs MR, Katz LM, et al. Survey of methods used to detect bacterial contamination of platelet products in the United States in 2011. Transfusion 2013;53:911-8. [Crossref] [PubMed]
- Kacker S, Katz LM, Ness PM, et al. Financial analysis of large-volume delayed sampling to reduce bacterial contamination of platelets. Transfusion 2020;60:997-1002. [Crossref] [PubMed]
- Mintz PD, Sanders JR. Outdate Reduction and Cost Savings with Rapid Testing for Seven-Day Platelet Storage. Ann Clin Lab Sci 2020;50:404-7. [PubMed]
Cite this article as: Jacobs MR. FDA guidance on bacterial contamination risk control strategies to enhance the safety and availability of platelets: advantages and limitations. Ann Blood 2021;6:18.


