
Analysis of hepatitis B virus (HBV) drug resistance variations in HBV carriers of Chinese blood donors
Introduction
Hepatitis B virus (HBV) infection is a global problem and about 120 million people in China carry HBV (1). At present, the treatment of chronic hepatitis B is mainly antiviral therapy (2), and the mainstream method is nucleoside drugs (3), but during the treatment period virus genes can mutate and lead to drug resistance. The aim of this study was to determine whether drug resistance genes were present in the HBV strains from blood donors.
Methods
Subjects
From April 1, 2014 to December 31, 2014, 201 blood samples, that were previously stored at −30 °C, were tested for positivity using two types of ELISAs (Lizhu HBsAg and Sorin Murex HBsAg version 3 assay kits).
Reagents
Enzyme-free kit: Zhuhai Lizhu HBsAg assay kit, batch numbers 2014010208, 2014020508, 2014051108, 2014081708; Sorin Murex HBsAg version 3 assay kit, batch numbers D188810, D224710, D224610; hepatitis B virus nucleic acid quantitative detection kit (Sun Yat-sen University Da’an Gene Co., Ltd.), batch numbers 2014008, 2014010, 2014016, 2014021; hepatitis B virus resistance gene mutation detection kit (Guangzhou Life Technologies Diagnostic Product Co., Ltd., PCR-sequencing—20 tests per box, the batch numbers of the detection reagents were 2014001 and 2014002.
Main instruments
Tecan Freedom Clinical 150/8 automatic sampler, FAME24/20 automatic enzyme-linked immunosorbent analyzer, BEPIII automatic enzyme-linked immunosorbent analyzer, Applied Biosystem 7500 real-time fluorescence PCR, Applied Biosystem 3500 XL Dx gene analyzer; analysis software: Applied Biosystem Data Collection and Sequencing Analysis software; data analysis software: Applied Biosystem Sequence Scanner Software version 2.0.
Experimental methods
First, two types of ELISA detection kits were used for HBsAg screening, and the positive samples were preserved. The preserved blood samples were subsequently analyzed by fluorescence quantitative PCR, operated according to the kit protocol, and nucleic acid extracted and primers added as required. The amplification conditions were as follows: 93 °C 2 min; 93 °C 45 s, 55 °C 60 s, 10 cycles, 93 °C 30 s, 55 °C 45 s, 30 cycles. YMDD probe [nt.717-731: antisense chain, 5'(6-Fam) ATCATCCATATARCTGA (HHQ1)-3']; YVDD probe [nt.747-731: antisense chain, 5'(Hex) ATCATCCACATARCTGA (HHQI)-3']; YIDD probe [nt.750-732: antisense chain, 5'(Cy5) CACATCATCAATATARCTG (HHQ3) 3']. If fluorescence quantitative PCR showed that the total amount of nucleic acid was higher than 1,000 IU/mL, Sanger sequencing was used.
An ABI 3500Dx gene sequencer was used to determine sequences and the denatured sequencing products were added to 96-well plates that matched the gene analyzer data. They were covered and the sample list was edited in the order of added samples According to the type of sequencer, Seq_std_BDTV3.1_ASSYXL_POP7 or Seq_std_BDTV3.1_ASSY_POP7 with IVD mark was selected for sequencing. When using the ABI gene analyzer, Data Collection and Sequencing Analysis software of ABI Company were used for data collection and analysis. The obtained target gene sequence was compared with the wild strain gene sequence to analyze whether there was any variation in the known drug resistance sites.
Results
Two hundred and one samples were detected positive by using two different ELISA kits, and among them 99 samples were detected positive by fluorescence quantitative PCR. In total, 45/99 samples had viral load greater than 1,000 IU/mL, which met the requirements for HBV DNA sequencing analysis (Table 1). HBV Pol region was amplified and amplicons (400 bp, amino acid position 143 to 280) were sequenced. Of 45 HBV samples 43 were genotype B and 2 were genotype C (4). The analysis of drug resistance sites was as follows (Table 2).
Table 1
| Sequence number | HBV-M model | Cases | ||||
|---|---|---|---|---|---|---|
| HBsAg | HBsAb | HBeAg | HBeAb | HBcAb | ||
| 1 | + | − | − | − | − | 3 |
| 2 | + | − | − | + | − | 1 |
| 3 | + | − | − | − | + | 5 |
| 4 | + | + | − | − | + | 1 |
| 5 | + | − | + | − | + | 1 |
| 6 | + | − | − | + | + | 30 |
| 7 | + | + | + | − | + | 2 |
| 8 | + | + | − | + | + | 2 |
| Total | 45 | |||||
Table 2
| Specimen No.*** | Amino acid site*/amino acid** | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 169/I | 173/V | 180/L | 181/A | 184/T | 194/A | 202/S | 204/M | 236/N | 250/M | |
| 1 | I | V | L | A | T | A | S | M | N | M |
| 2 | ? | V | ? | A | T | A | S | M | ? | M |
| 3 | I | V | L | A | T | A | S | M | N | M |
| 4 | I | V | L | A | T | A | S | M | N | M |
| 5 | I | V | L | A | T | A | S | M | N | M |
| 6 | I | V | L | A | T | A | S | M | N | M |
| 7 | I | V | L | A | T | A | S | M | N | M |
| 8 | I | V | L | A | T | A | S | M | N | M |
| 9 | I | V | L | A | T | A | S | M | N | M |
| 10 | I | V | L | A | T | A | S | M | N | M |
| 11 | I | V | L | A | T | A | S | M | N | M |
| 12 | I | V | L | A | T | A | S | M | N | M |
| 13 | I | V | L | A | T | A | S | M | N | M |
| 14 | I | V | L | A | T | A | S | M | N | M |
| 15 | I | V | L | A | T | A | S | M | N | M |
| 16 | I | V | L | A | T | A | S | M | N | M |
| 17 | I | V | L | A | T | A | S | M | N | M |
| 18 | I | V | L | A | T | A | S | M | N | M |
| 19 | I | V | L | A | T | A | S | M | N | M |
| 20 | I | V | L | A | T | A | S | M | N | M |
| 21 | I | V | L | A | T | A | S | M | N | M |
| 22 | I | V | L | A | T | A | S | M | N | M |
| 23 | I | V | L | A | T | A | S | M | N | M |
| 24 | I | V | L | A | T | A | S | M | N | M |
| 25 | I | V | L | A | T | A | S | M | N | M |
| 26 | I | V | L | A | T | A | S | M | N | M |
| 27 | I | V | L | A | T | A | S | M | N | M |
| 28 | I | V | L | A | T | A | S | M | N | M |
| 29 | I | V | L | A | T | A | S | M | N | M |
| 30 | I | V | L | A | T | A | S | M | N | M |
| 31 | I | V | L | A | T | A | S | M | N | M |
| 32 | I | V | L | A | T | A | S | M | N | M |
| 33 | I | V | L | A | T | A | S | M | N | M |
| 34 | I | V | L | A | T | A | S | M | N | M |
| 35 | I | V | L | A | T | A | S | M | N | M |
| 36 | I | V | L | A | T | A | S | M | N | M |
| 37 | I | ? | L | A | T | A | S | M | N | M |
| 38 | I | V | L | A | T | A | S | M | N | M |
| 39 | I | V | L | A | T | A | S | M | N | M |
| 40 | I | V | L | A | T | A | S | M | N | M |
| 41 | I | V | L | A | T | A | S | M | N | M |
| 42 | I | V | L | A | T | A | S | M | N | M |
| 43 | I | V | L | A | T | A | S | M | N | M |
| 44 | ? | ? | L | A | T | ? | S | M | N | M |
| 45 | ? | ? | L | A | T | ? | S | M | N | M |
*, the mutation sites relative to drug resistance; **, the corresponding amino acid sites of the wild type of HBV; ***, the amino acid sites detected in HBV strains from blood donors. “?” indicates that the sequencing results are unable to identify the amino acid by sequencing reason, so that the corresponding amino acid site could not be unequivocally determined.
Discussion
HBV is a DNA virus, about 3.2 kb long, in the form of incomplete double-stranded circular DNA. The genome structure of HBV is particularly precise and concentrated and there are many overlapping gene sequences. Four identified open reading frames have been identified in the HBV genome, which are termed the S, C, P and X regions, respectively. The core-shell (C) and envelope (S) protein, HBV polymerase and a protein X (5) that seem to be related to the expression of the virus gene are encoded respectively in these virus regions. At present, nucleoside drugs mainly inhibit virus replication by inhibiting the DNA polymerase activity of HBV. All known nucleotide resistance sites occur in the functional region of reverse transcriptase (6), and the naming of drug resistance sites (7) has been unified internationally, starting from the first amino acid upstream of the reverse transcriptase region. Rt1–344 (the reverse transcriptase domain of various genotypes of HBV is comprised of 344 amino acids) was used to name and locate the mutation sites related to nucleotides. Life technologies reagents mainly detect HBV pol region. Through sequencing reaction detection, more than a dozen known drug resistance mutation sites in the RT reverse transcription region can be identified at the same time, including all the corresponding drug resistance sites that have been reported in the published literature. The sequencing length of hepatitis B in this region is generally greater than 400 bases (amino acid position 143 to 280) as shown in Figure 1 (8).
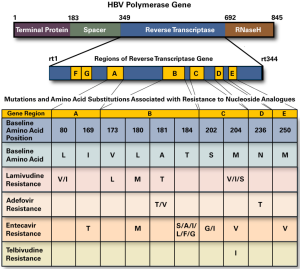
Table 1 shows all the combinations of HBV-M patterns of 45 samples that could be sequenced. The main combination was 30 samples with all HBsAg, HBeAb, HBcAb positive. Only two cases of ALT were greater and the others were normal, which accorded with the characteristics of HBV carriers in the asymptomatic population. As shown in Table 2, the obtained sequences could be used to analyze 10 sites, such as 169, 173, 180, 181, 184, 194, 202, 204, 236, 250, etc., except for 1 site in 1 case and 3 sites in 3 cases, which could not be clearly determined. The other case clearly determined the corresponding amino acids, but all were not mutated in comparison with wild type HBV strains.
Previously it was believed that drugs induced mutations in HBV, resulting in drug resistance. Subsequent studies have shown that due to the lack of a correction function of the RNA enzyme and reverse transcriptase (9), the probability of variation in virus replication is quite high, and that HBV-DNA can mutate naturally in the process of chronic persistent infection. It can also mutate during immune pressure, even under the induction of various antiviral drugs (10). Wild type strains and naturally mutated drug-resistant strains coexist before drug use. Drugs only play a screening role after treatment and sensitive wild strains are eliminated and mutants become dominant strains. Many scholars believe that there are naturally mutated drug-resistant strains. Chen et al. detected HBV-associated HCC in 110 patients and 1,079 patients with chronic HBV infection (including hepatitis B carriers, chronic hepatitis B and hepatitis B cirrhosis); all these patients did not receive any nucleoside (acid) or analogues treatment. There were 16 and 46 cases of natural variation of HBV YMDD in these two groups. The results showed that YMDD natural variation combined with the type C gene was an independent risk factor for HBV-associated HCC (11). Chen et al. detected 14 cases of natural variation of YMDD in serum samples of 152 cases of chronic hepatitis B and 18 cases of hepatitis B sclerosis without treatment with Chinese or western medicine. It was considered that the natural variation of YMDD of HBV could occur in the process of chronicity of infection in patients, co-existing with wild strains (12). Clinicians call this untreated drug resistance site in patients infected with HBV as a pre-existing drug resistance (13).
At present, the analysis of drug resistance of hepatitis B in clinical mainly aims for the hospitalized patients, and less for blood donors. Most of the small volume of samples from blood donors in China are used for detection of HBV, and few for analysis of drug resistance sites. The objective of the present study was to analyze HBV drug resistance in blood donors who were not received drug treatment. None of these 45 HBV strains had detectable mutation at drug resistance sites, which indicated that the prevalent HBV strains in this region were wild type of HBV. At the same time, the test results do not prove the view that there is a natural variation in drug resistance sites. It is hopeful that with further development in detection technology, there will be a large number of blood samples from asymptomatic HBV carriers for epidemiological investigation on HBV drug resistance.
Acknowledgments
Funding: Financial support from Guangzhou Life Technologies Diagnostic Product Co., Ltd.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aob.2019.06.03). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. According to the Chinese national standard “Whole blood and component selection requirements” (GB18467-2011), it is necessary to detect hepatitis B virus in blood donors. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Before blood donation, informed consent and signature has been obtained from every blood donor. This article does not apply for ethical review.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gu J, Zhuo C, Li Yan. Research Progress of Anti-Hepatitis B virus drugs. Modern Practical Medicine 2007;34:110-4.
- Jiang P, Zeng W. Advances in drug resistance of lamivudine in the treatment of chronic hepatitis B. Medical Information 2011;4:345-6.
- Zhang S, Yu W, Zhou D, et al. Study on hepatitis B virus resistance genotype in patients with chronic hepatitis B. International Journal of Laboratory Medicine 201;32:1797-9.
- Xiao Z, Lin S, Gan X. Research for HBV Genotype and Mutation in Blood Donation Population in a Given Area. J Clin Transfus Lab Med 2016;18:444-7.
- Tong L, Tong L. Research Progress and Prospect of Hepatitis B Vaccine. Modern Practical Medicine 2007;19:78-81.
- Shelly X. Progress in the study of drug resistance of hepatitis B virus. Liver 2016;11:273-5.
- Stuyver LJ, Locarnini SA, Lok A, et al. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology 2001;33:751-7. [Crossref] [PubMed]
- Lok AS, Zoulim F, Locarnini S, et al. Antiviral drug-resistant HBV: standardization of nomenclature and assays and recommendations for management. Hepatology 2007;46:254-65. [Crossref] [PubMed]
- Beck J, Nassal M. Hepatitis B virus replication. World J Gastroenterol 2007;13:48-64. [Crossref] [PubMed]
- Gutfreund KS, Williams M, George R, et al. Genotypic succession of mutations of the hepatitis B virus polymerase associated with lamivudine resistance. J Hepatol 2000;33:469-75. [Crossref] [PubMed]
- Chen X, Yang J, Zhang H, et al. Relationship between hepatitis B virus YMDD natural mutation combined with genotype B and C and hepatitis B virus-associated hepatocellular carcinoma. West China Medicine 2015;30:1650-3.
- Chen N, Zhao S, Liu X, et al. Sequence variation of HBV P gene YMDD gene without antiviral therapy and its relationship with genotype. Chin J Clinicians 2008;2:1123-7. (Electronic Version).
- Luo X, Han F, Zhang L. Study on pre-stored drug resistance of hepatitis B virus reverse transcriptase region. Chinese Journal of Applied Medicine 2015;35:786-9.
Cite this article as: Xiao Z, Gan X, Lin S, Zheng Z. Analysis of hepatitis B virus (HBV) drug resistance variations in HBV carriers of Chinese blood donors. Ann Blood 2019;4:11.


